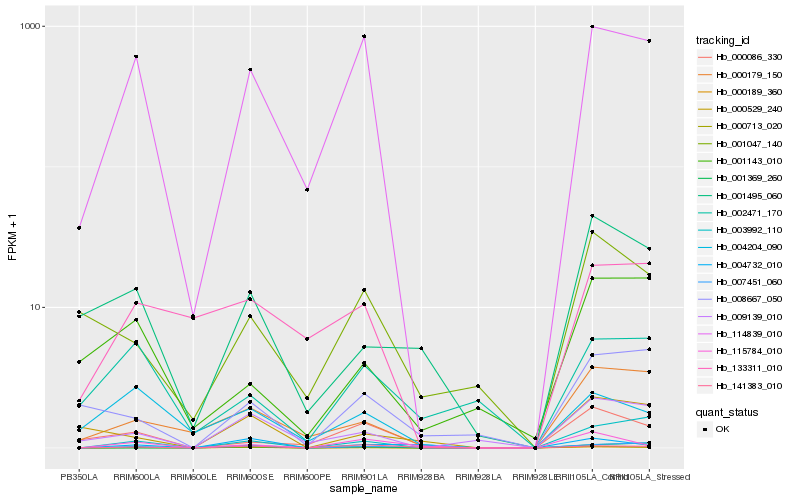
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_360 |
0.0 |
- |
- |
Calcium-transporting ATPase 12, plasma membrane-type [Morus notabilis] |
| 2 |
Hb_114839_010 |
0.1942450438 |
- |
- |
- |
| 3 |
Hb_007451_060 |
0.2287383038 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 4 |
Hb_003992_110 |
0.2317273104 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 5 |
Hb_009139_010 |
0.2327452199 |
- |
- |
putative Phospholipid-Diacylglycerol acyltransferase [Elaeis guineensis] |
| 6 |
Hb_000713_020 |
0.2350344435 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000179_150 |
0.2617002748 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 8 |
Hb_141383_010 |
0.2868697111 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 9 |
Hb_004732_010 |
0.2933030004 |
- |
- |
- |
| 10 |
Hb_000529_240 |
0.2978084285 |
- |
- |
- |
| 11 |
Hb_001495_060 |
0.305294295 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 12 |
Hb_133311_010 |
0.307538667 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 13 |
Hb_008667_050 |
0.3154371442 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 14 |
Hb_004204_090 |
0.3163454139 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 15 |
Hb_001369_260 |
0.3185029515 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_000086_330 |
0.3346310168 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_115784_010 |
0.334922075 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 18 |
Hb_001143_010 |
0.3372781173 |
- |
- |
- |
| 19 |
Hb_002471_170 |
0.339414759 |
- |
- |
- |
| 20 |
Hb_001047_140 |
0.3405443732 |
- |
- |
- |