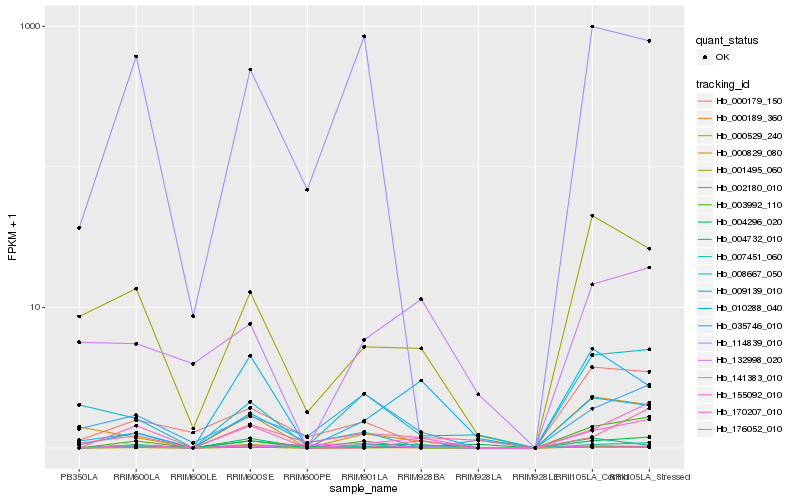
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007451_060 |
0.0 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 2 |
Hb_000189_360 |
0.2287383038 |
- |
- |
Calcium-transporting ATPase 12, plasma membrane-type [Morus notabilis] |
| 3 |
Hb_003992_110 |
0.2368977566 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 4 |
Hb_170207_010 |
0.2519561194 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 5 |
Hb_141383_010 |
0.2757839014 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 6 |
Hb_000529_240 |
0.2776993847 |
- |
- |
- |
| 7 |
Hb_000829_080 |
0.2824113453 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 8 |
Hb_000179_150 |
0.2831443941 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 9 |
Hb_004732_010 |
0.2908714109 |
- |
- |
- |
| 10 |
Hb_001495_060 |
0.2922350452 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 11 |
Hb_008667_050 |
0.2924851657 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 12 |
Hb_176052_010 |
0.2958913809 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_155092_010 |
0.2962626115 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 14 |
Hb_004296_020 |
0.2980267496 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_035746_010 |
0.3037199866 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 16 |
Hb_009139_010 |
0.3039487856 |
- |
- |
putative Phospholipid-Diacylglycerol acyltransferase [Elaeis guineensis] |
| 17 |
Hb_132998_020 |
0.3052748883 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002180_010 |
0.3058237504 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_010288_040 |
0.3064595172 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 20 |
Hb_114839_010 |
0.3082895494 |
- |
- |
- |