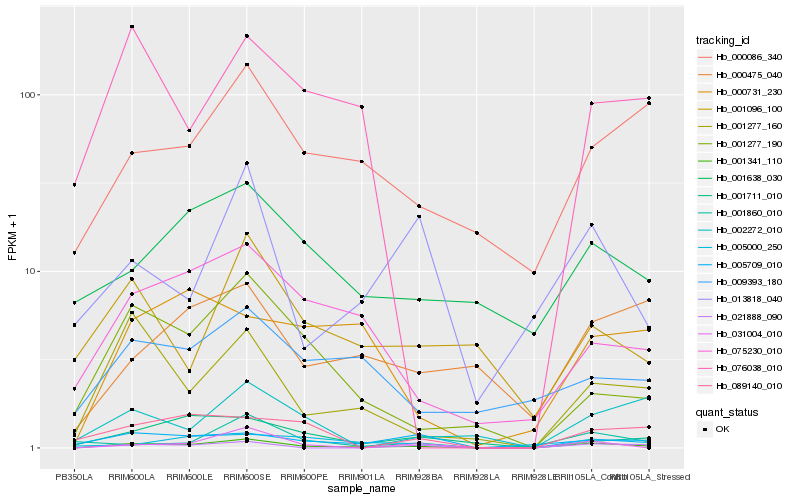
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001341_110 |
0.0 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 2 |
Hb_001277_160 |
0.2542491302 |
- |
- |
hypothetical protein MIMGU_mgv1a019742mg, partial [Erythranthe guttata] |
| 3 |
Hb_021888_090 |
0.2555337763 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 isoform X1 [Malus domestica] |
| 4 |
Hb_075230_010 |
0.2764963653 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 5 |
Hb_001277_190 |
0.2784354518 |
- |
- |
hypothetical protein CICLE_v10005534mg [Citrus clementina] |
| 6 |
Hb_000086_340 |
0.2793209134 |
- |
- |
PREDICTED: uncharacterized protein LOC100253191 [Vitis vinifera] |
| 7 |
Hb_005000_250 |
0.2857869624 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 4, mitochondrial [Vitis vinifera] |
| 8 |
Hb_031004_010 |
0.291795711 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 9 |
Hb_005709_010 |
0.293099558 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000475_040 |
0.2991244989 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001711_010 |
0.2992905081 |
- |
- |
- |
| 12 |
Hb_009393_180 |
0.3001410682 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 13 |
Hb_089140_010 |
0.3009107185 |
- |
- |
- |
| 14 |
Hb_002272_010 |
0.300942085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_076038_010 |
0.3009685984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001638_030 |
0.3030114074 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 17 |
Hb_013818_040 |
0.3036677137 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 18 |
Hb_001860_010 |
0.3041627462 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 19 |
Hb_001096_100 |
0.3045862218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000731_230 |
0.3052498423 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |