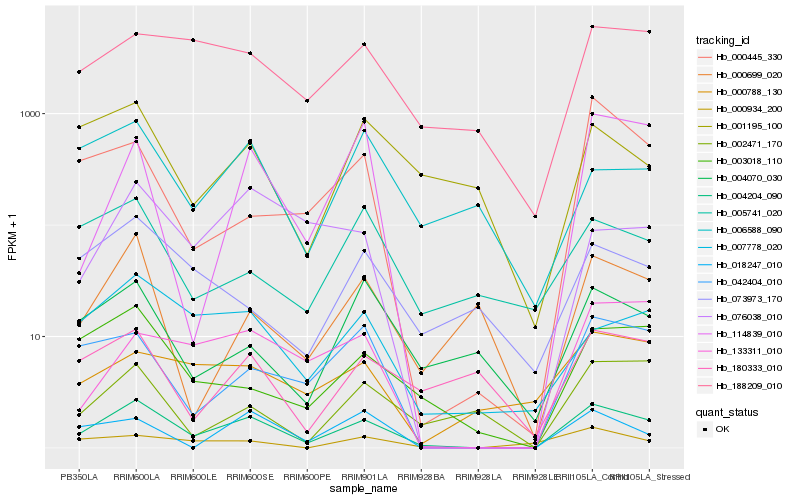
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004204_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 2 |
Hb_042404_010 |
0.2238430879 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 3 |
Hb_003018_110 |
0.2283523652 |
- |
- |
- |
| 4 |
Hb_007778_020 |
0.2300557113 |
- |
- |
- |
| 5 |
Hb_188209_010 |
0.2320111535 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_114839_010 |
0.2487559077 |
- |
- |
- |
| 7 |
Hb_133311_010 |
0.2523965202 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 8 |
Hb_180333_010 |
0.2550823138 |
- |
- |
- |
| 9 |
Hb_018247_010 |
0.2555552867 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 10 |
Hb_002471_170 |
0.2565426899 |
- |
- |
- |
| 11 |
Hb_000699_020 |
0.257477201 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 12 |
Hb_004070_030 |
0.2585412654 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000788_130 |
0.259094195 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 14 |
Hb_076038_010 |
0.260232625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000934_200 |
0.2609004821 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 16 |
Hb_001195_100 |
0.2632602305 |
- |
- |
PREDICTED: uncharacterized protein LOC105633812 [Jatropha curcas] |
| 17 |
Hb_000445_330 |
0.2643966219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_073973_170 |
0.2663624967 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 19 |
Hb_005741_020 |
0.2678541779 |
- |
- |
PREDICTED: putative golgin subfamily A member 6-like protein 6 [Jatropha curcas] |
| 20 |
Hb_006588_090 |
0.2693483194 |
- |
- |
PREDICTED: uncharacterized protein At4g28440 [Jatropha curcas] |