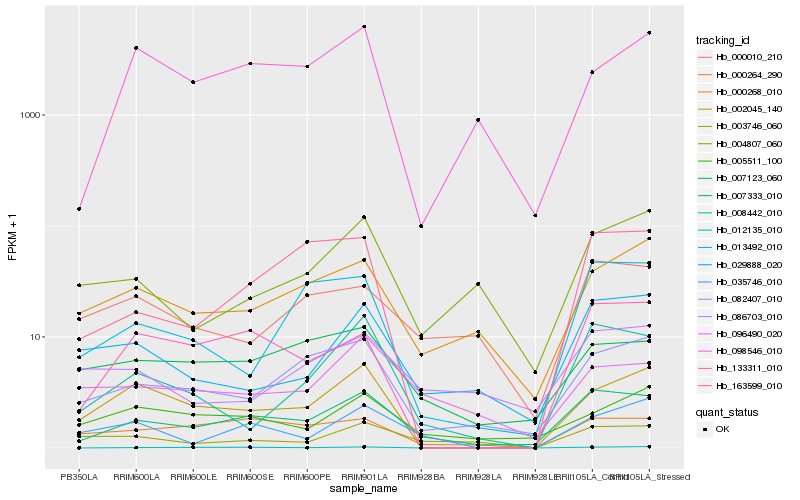
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007333_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_082407_010 |
0.1718472911 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_000268_010 |
0.1961533979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007123_060 |
0.1996213571 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 5 |
Hb_008442_010 |
0.2010876802 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 6 |
Hb_000264_290 |
0.2058986604 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 7 |
Hb_133311_010 |
0.2080697511 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 8 |
Hb_029888_020 |
0.2092616927 |
- |
- |
hypothetical protein PHAVU_007G268100g [Phaseolus vulgaris] |
| 9 |
Hb_013492_010 |
0.2117171573 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 10 |
Hb_096490_020 |
0.2129589188 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 11 |
Hb_000010_210 |
0.2138492194 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 12 |
Hb_163599_010 |
0.2150700704 |
- |
- |
hypothetical protein RCOM_1579620 [Ricinus communis] |
| 13 |
Hb_012135_010 |
0.2164076594 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 14 |
Hb_004807_060 |
0.2185931874 |
- |
- |
- |
| 15 |
Hb_005511_100 |
0.2190884152 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 16 |
Hb_086703_010 |
0.2205554641 |
- |
- |
PREDICTED: F-box protein At5g52880 [Jatropha curcas] |
| 17 |
Hb_003746_060 |
0.2212288995 |
- |
- |
PREDICTED: DTW domain-containing protein 2 [Jatropha curcas] |
| 18 |
Hb_035746_010 |
0.2233638809 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 19 |
Hb_098546_010 |
0.2255135078 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 20 |
Hb_002045_140 |
0.2265445663 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |