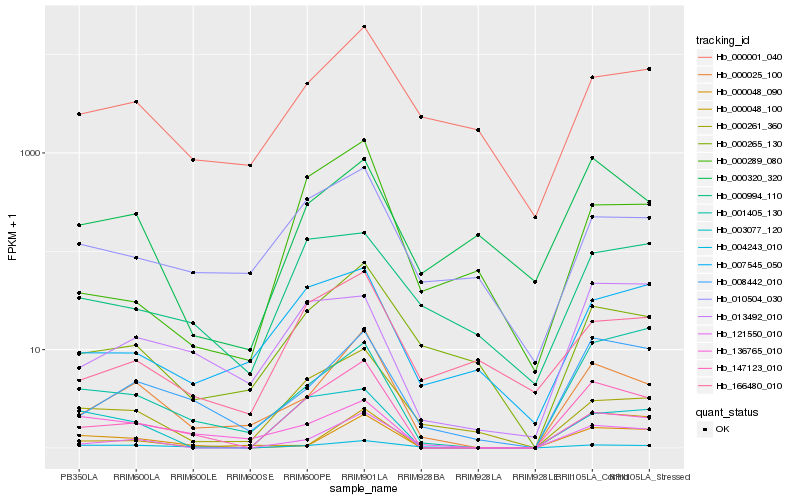
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147123_010 |
0.0 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000025_100 |
0.1877241613 |
- |
- |
- |
| 3 |
Hb_121550_010 |
0.1891410648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008442_010 |
0.2000756103 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 5 |
Hb_007545_050 |
0.2409593308 |
- |
- |
- |
| 6 |
Hb_000048_090 |
0.2416100565 |
transcription factor |
TF Family: SET |
hypothetical protein JCGZ_06559 [Jatropha curcas] |
| 7 |
Hb_000265_130 |
0.2454207055 |
- |
- |
- |
| 8 |
Hb_010504_030 |
0.2475557263 |
- |
- |
calmodulin homolog {EST} [Brassica napus, Naehan, root, Peptide Partial, 53 aa] |
| 9 |
Hb_003077_120 |
0.2480244629 |
- |
- |
- |
| 10 |
Hb_013492_010 |
0.2495101903 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 11 |
Hb_000261_360 |
0.2495428574 |
- |
- |
unknown [Lotus japonicus] |
| 12 |
Hb_166480_010 |
0.2499554237 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 13 |
Hb_004243_010 |
0.2502387094 |
- |
- |
hypothetical protein JCGZ_01580 [Jatropha curcas] |
| 14 |
Hb_000289_080 |
0.2510360323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_136765_010 |
0.2554107535 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001405_130 |
0.2559319198 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 17 |
Hb_000001_040 |
0.2576599828 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_000320_320 |
0.258062605 |
- |
- |
hypothetical protein POPTR_0004s01360g [Populus trichocarpa] |
| 19 |
Hb_000994_110 |
0.2626486177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000048_100 |
0.2654896663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |