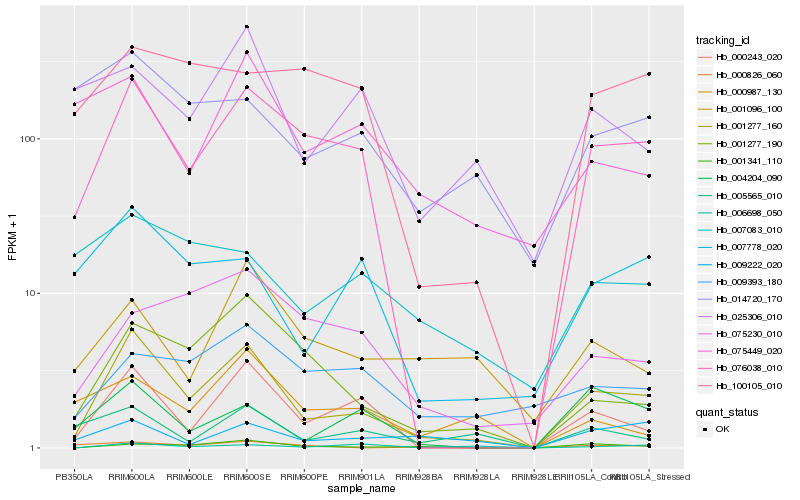
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_160 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a019742mg, partial [Erythranthe guttata] |
| 2 |
Hb_076038_010 |
0.196281547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000243_020 |
0.2147847483 |
- |
- |
- |
| 4 |
Hb_001277_190 |
0.2413507385 |
- |
- |
hypothetical protein CICLE_v10005534mg [Citrus clementina] |
| 5 |
Hb_001341_110 |
0.2542491302 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 6 |
Hb_007778_020 |
0.2695706134 |
- |
- |
- |
| 7 |
Hb_004204_090 |
0.2704593607 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 8 |
Hb_005565_010 |
0.2780878579 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 9 |
Hb_001096_100 |
0.2881598672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009222_020 |
0.2890766925 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 11 |
Hb_009393_180 |
0.2907856698 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_000826_060 |
0.293739263 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 13 |
Hb_025306_010 |
0.2941429682 |
transcription factor |
TF Family: MYB-related |
syringolide-induced protein 1-3-1B [Populus trichocarpa] |
| 14 |
Hb_075230_010 |
0.2958622636 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 15 |
Hb_075449_020 |
0.2977609021 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 16 |
Hb_000987_130 |
0.2999328025 |
- |
- |
- |
| 17 |
Hb_100105_010 |
0.3019131976 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 18 |
Hb_014720_170 |
0.302549649 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 19 |
Hb_006698_050 |
0.3027415375 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 20 |
Hb_007083_010 |
0.3030389571 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 13-like, partial [Nelumbo nucifera] |