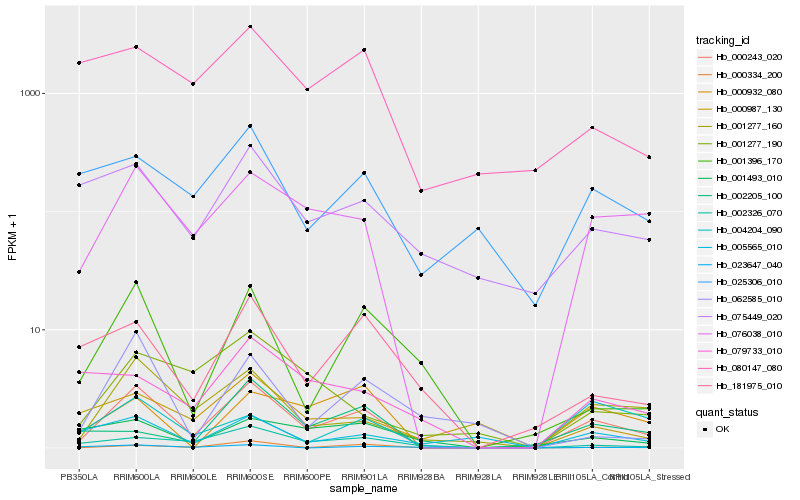
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_076038_010 |
0.1895376152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001277_160 |
0.2147847483 |
- |
- |
hypothetical protein MIMGU_mgv1a019742mg, partial [Erythranthe guttata] |
| 4 |
Hb_000334_200 |
0.22670561 |
desease resistance |
Gene Name: RVT_1 |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 5 |
Hb_002326_070 |
0.2567342234 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000932_080 |
0.2616576491 |
- |
- |
- |
| 7 |
Hb_001396_170 |
0.2752888943 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_181975_010 |
0.2766923975 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 9 |
Hb_025306_010 |
0.2880397741 |
transcription factor |
TF Family: MYB-related |
syringolide-induced protein 1-3-1B [Populus trichocarpa] |
| 10 |
Hb_001277_190 |
0.2889237514 |
- |
- |
hypothetical protein CICLE_v10005534mg [Citrus clementina] |
| 11 |
Hb_005565_010 |
0.2890873474 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 12 |
Hb_075449_020 |
0.2918149209 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 13 |
Hb_001493_010 |
0.2953627925 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 14 |
Hb_000987_130 |
0.2953675248 |
- |
- |
- |
| 15 |
Hb_004204_090 |
0.2961250997 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 16 |
Hb_023647_040 |
0.2978904388 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 17 |
Hb_080147_080 |
0.2996241468 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 18 |
Hb_062585_010 |
0.299975885 |
- |
- |
hypothetical protein JCGZ_19532 [Jatropha curcas] |
| 19 |
Hb_002205_100 |
0.3002823089 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_079733_010 |
0.3076181977 |
- |
- |
- |