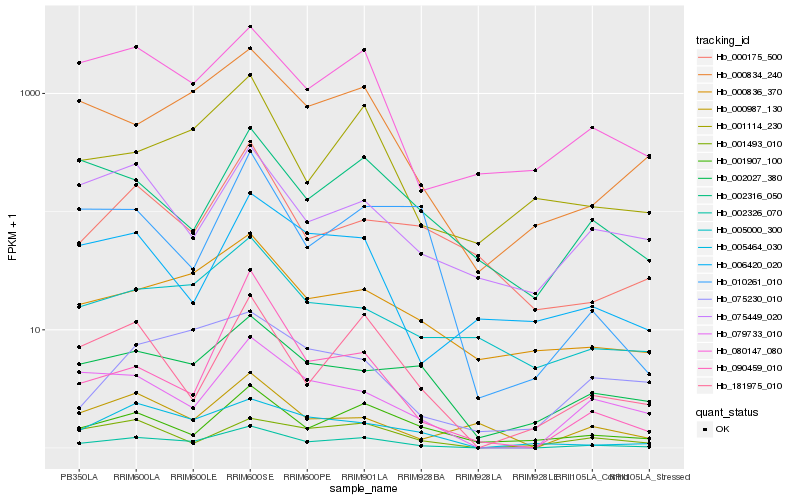
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002326_070 |
0.0 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_080147_080 |
0.1853192885 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 3 |
Hb_079733_010 |
0.1854632965 |
- |
- |
- |
| 4 |
Hb_181975_010 |
0.1871385183 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |
| 5 |
Hb_005464_030 |
0.1925150121 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000834_240 |
0.1926255622 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 7 |
Hb_001907_100 |
0.1968436754 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 8 |
Hb_002027_380 |
0.202718281 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 9 |
Hb_000836_370 |
0.2069500466 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001114_230 |
0.207899605 |
- |
- |
- |
| 11 |
Hb_000987_130 |
0.2172223803 |
- |
- |
- |
| 12 |
Hb_005000_300 |
0.2206552366 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 13 |
Hb_002316_050 |
0.2248719796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006420_020 |
0.2250614596 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 15 |
Hb_001493_010 |
0.2261411048 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 16 |
Hb_075449_020 |
0.226292466 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 17 |
Hb_075230_010 |
0.2264906493 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 18 |
Hb_090459_010 |
0.2267147901 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 19 |
Hb_000175_500 |
0.2267917905 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 20 |
Hb_010261_010 |
0.2302459447 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |