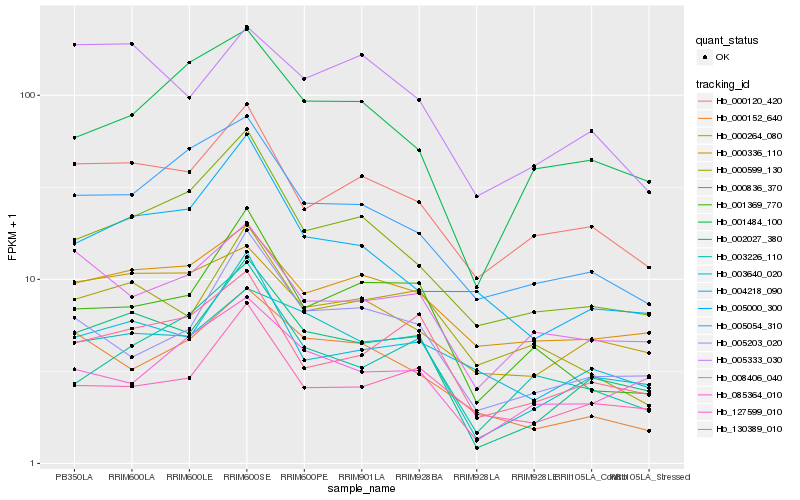
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_380 |
0.0 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 2 |
Hb_003640_020 |
0.1074947686 |
- |
- |
pelota, putative [Ricinus communis] |
| 3 |
Hb_130389_010 |
0.1265090619 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 4 |
Hb_000120_420 |
0.1357260749 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_000836_370 |
0.1373114615 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005203_020 |
0.1439765464 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 7 |
Hb_001369_770 |
0.1483654224 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 8 |
Hb_000336_110 |
0.1484981559 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_005054_310 |
0.1491023556 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 10 |
Hb_004218_090 |
0.150066688 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_127599_010 |
0.1503742327 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 12 |
Hb_000599_130 |
0.1512902326 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 13 |
Hb_000264_080 |
0.1521944958 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 14 |
Hb_008406_040 |
0.1543665769 |
- |
- |
- |
| 15 |
Hb_001484_100 |
0.1552725077 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 16 |
Hb_005000_300 |
0.1564610078 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 17 |
Hb_085364_010 |
0.157105174 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 18 |
Hb_005333_030 |
0.1595350503 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 19 |
Hb_003226_110 |
0.1606408758 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 20 |
Hb_000152_640 |
0.1608384425 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |