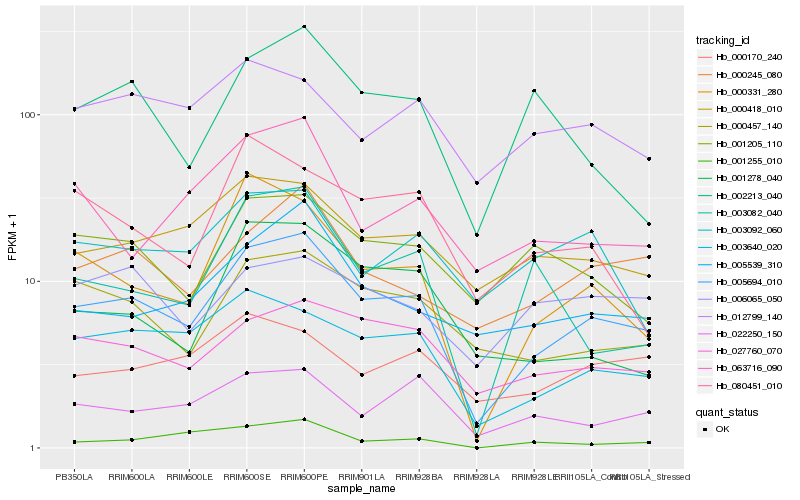
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_010 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_003640_020 |
0.1280259541 |
- |
- |
pelota, putative [Ricinus communis] |
| 3 |
Hb_000331_280 |
0.1431878149 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_027760_070 |
0.1446362401 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 5 |
Hb_000418_010 |
0.1486208304 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 6 |
Hb_000245_080 |
0.1500585632 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 7 |
Hb_000457_140 |
0.1507203939 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 8 |
Hb_005539_310 |
0.1542095725 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 9 |
Hb_022250_150 |
0.1566875244 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 10 |
Hb_063716_090 |
0.1636896556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000170_240 |
0.1676049344 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 12 |
Hb_001205_110 |
0.1692475118 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 13 |
Hb_001278_040 |
0.1717256055 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_012799_140 |
0.1721861283 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_003092_060 |
0.1727461576 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_002213_040 |
0.1738416988 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 17 |
Hb_003082_040 |
0.1748108589 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 18 |
Hb_001255_010 |
0.1752487093 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Tarenaya hassleriana] |
| 19 |
Hb_080451_010 |
0.1754857247 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 20 |
Hb_006065_050 |
0.1785995542 |
transcription factor |
TF Family: AP2 |
PREDICTED: floral homeotic protein APETALA 2 isoform X1 [Jatropha curcas] |