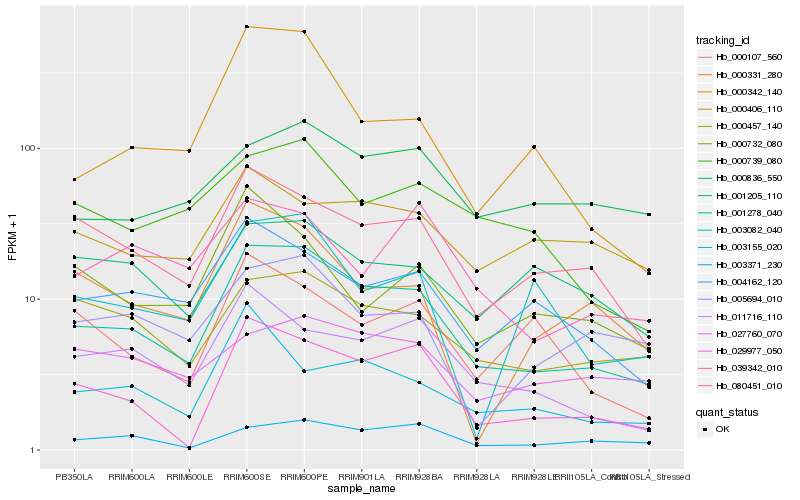
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001278_040 |
0.0 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_080451_010 |
0.1378503499 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 3 |
Hb_029977_050 |
0.1440304093 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000457_140 |
0.1500841379 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 5 |
Hb_004162_120 |
0.1546522959 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 6 |
Hb_011716_110 |
0.1546647677 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 7 |
Hb_003371_230 |
0.1586844244 |
- |
- |
lactase-like b precursor [Scleropages formosus] |
| 8 |
Hb_000342_140 |
0.1668674975 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 9 |
Hb_000739_080 |
0.1683771194 |
- |
- |
PREDICTED: 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase [Jatropha curcas] |
| 10 |
Hb_005694_010 |
0.1717256055 |
transcription factor |
TF Family: C2C2-Dof |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 11 |
Hb_000836_550 |
0.1781952324 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 12 |
Hb_000107_560 |
0.1793108243 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 13 |
Hb_000331_280 |
0.1794973145 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 14 |
Hb_003082_040 |
0.1818261911 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 15 |
Hb_027760_070 |
0.182378333 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 16 |
Hb_039342_010 |
0.1834102133 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 17 |
Hb_001205_110 |
0.1841954716 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 18 |
Hb_003155_020 |
0.1865522835 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 19 |
Hb_000406_110 |
0.1871897566 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 20 |
Hb_000732_080 |
0.1880240675 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |