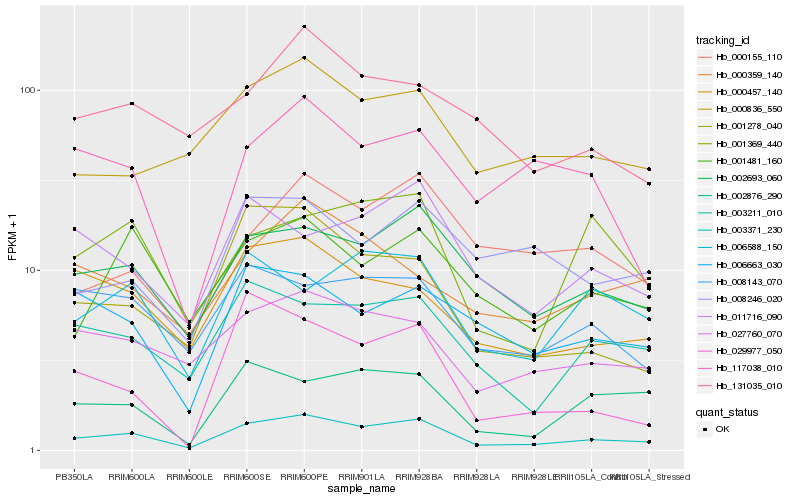
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_230 |
0.0 |
- |
- |
lactase-like b precursor [Scleropages formosus] |
| 2 |
Hb_002693_060 |
0.1365560434 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_027760_070 |
0.1538697306 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 4 |
Hb_003211_010 |
0.1545526095 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 5 |
Hb_001278_040 |
0.1586844244 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000457_140 |
0.1618583427 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 7 |
Hb_001481_160 |
0.1640849904 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 8 |
Hb_002876_290 |
0.1646737237 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 9 |
Hb_000836_550 |
0.1677253145 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 10 |
Hb_008143_070 |
0.1689972212 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 11 |
Hb_006588_150 |
0.170198559 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 12 |
Hb_006663_030 |
0.1707067806 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_001369_440 |
0.1736584724 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 14 |
Hb_000155_110 |
0.1738440676 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 15 |
Hb_011716_090 |
0.1744066718 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 16 |
Hb_117038_010 |
0.1783594079 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 17 |
Hb_029977_050 |
0.1788657983 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_131035_010 |
0.1790995126 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000359_140 |
0.1793550543 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 20 |
Hb_008246_020 |
0.1794383239 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |