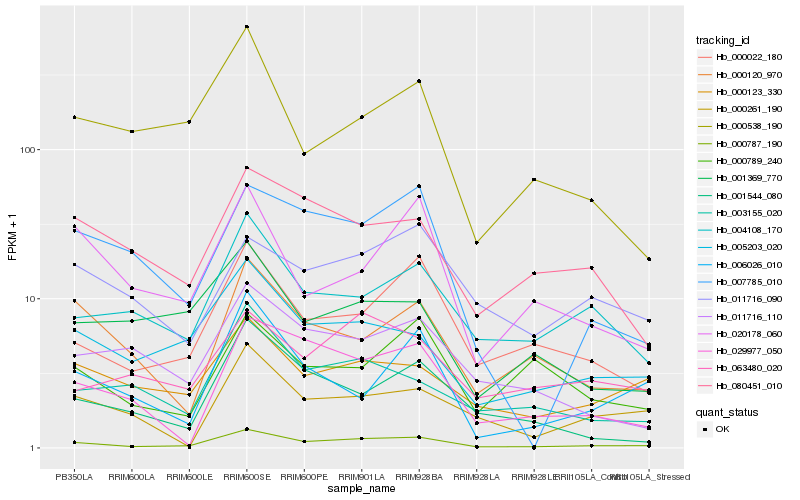
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_190 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 15-like [Jatropha curcas] |
| 2 |
Hb_000022_180 |
0.1548427202 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005203_020 |
0.1638009572 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 4 |
Hb_004108_170 |
0.1759433221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000123_330 |
0.1783212461 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 6 |
Hb_029977_050 |
0.1792495766 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003155_020 |
0.1856738358 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 8 |
Hb_000120_970 |
0.18774705 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_063480_020 |
0.1910870644 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_001544_080 |
0.1915811611 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 11 |
Hb_006026_010 |
0.1929383887 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 3-like [Jatropha curcas] |
| 12 |
Hb_080451_010 |
0.1945852012 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 13 |
Hb_011716_110 |
0.1948146415 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 14 |
Hb_000261_190 |
0.195704792 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 15 |
Hb_007785_010 |
0.1984191031 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 16 |
Hb_000789_240 |
0.198448638 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 17 |
Hb_020178_060 |
0.2001136494 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 18 |
Hb_000538_190 |
0.2003374813 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 19 |
Hb_001369_770 |
0.2013298336 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 20 |
Hb_011716_090 |
0.2034465349 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |