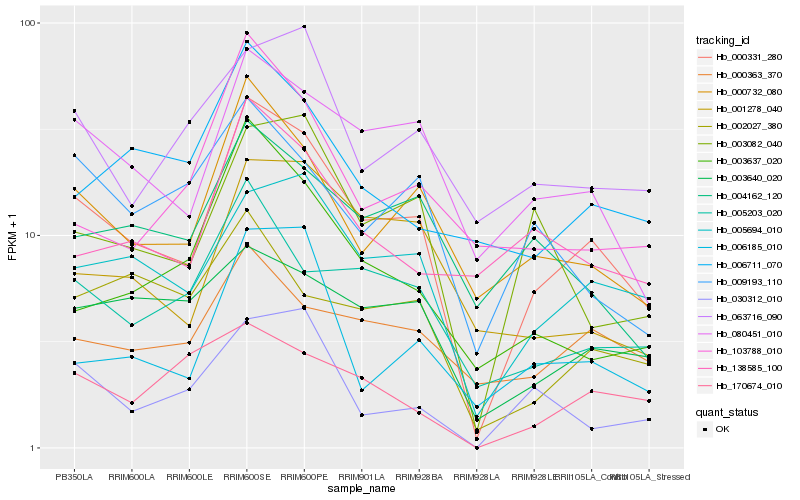
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_280 |
0.0 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 2 |
Hb_005694_010 |
0.1431878149 |
transcription factor |
TF Family: C2C2-Dof |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_000732_080 |
0.1442025388 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 4 |
Hb_080451_010 |
0.1453179798 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 5 |
Hb_005203_020 |
0.1705736379 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 6 |
Hb_003082_040 |
0.1712522193 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 7 |
Hb_001278_040 |
0.1794973145 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003640_020 |
0.180295739 |
- |
- |
pelota, putative [Ricinus communis] |
| 9 |
Hb_004162_120 |
0.1809500218 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 10 |
Hb_006185_010 |
0.1822656977 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 11 |
Hb_000363_370 |
0.1848630593 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 12 |
Hb_003637_020 |
0.1881961068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009193_110 |
0.1900237468 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_063716_090 |
0.1904768655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006711_070 |
0.1915105997 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 16 |
Hb_138585_100 |
0.1922992531 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 17 |
Hb_103788_010 |
0.1929573297 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_030312_010 |
0.1937019384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002027_380 |
0.1938217073 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 20 |
Hb_170674_010 |
0.1994266211 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |