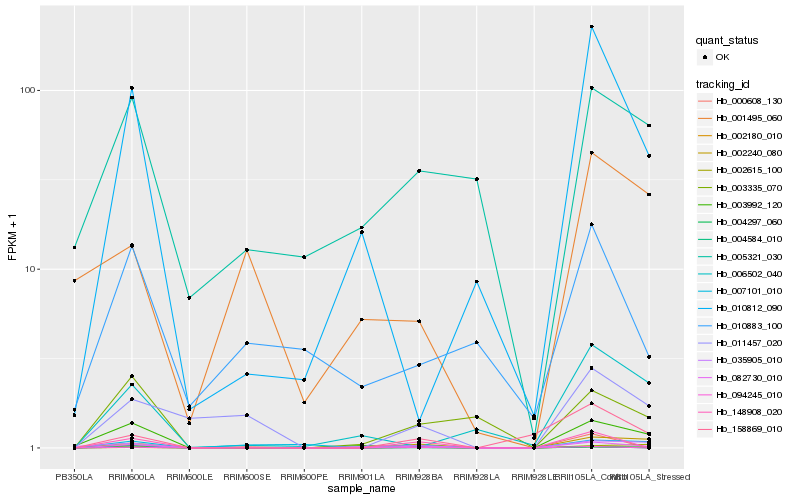
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_082730_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 2 |
Hb_002240_080 |
0.25660153 |
- |
- |
- |
| 3 |
Hb_011457_020 |
0.266366693 |
- |
- |
PREDICTED: uncharacterized protein LOC105649960 [Jatropha curcas] |
| 4 |
Hb_094245_010 |
0.288206148 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 5 |
Hb_004297_060 |
0.3020659736 |
- |
- |
Cucumisin protein [Theobroma cacao] |
| 6 |
Hb_148908_020 |
0.3023692857 |
- |
- |
- |
| 7 |
Hb_002180_010 |
0.3042691011 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_007101_010 |
0.3044306402 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 9 |
Hb_003992_120 |
0.323339019 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 10 |
Hb_002615_100 |
0.3372465392 |
- |
- |
- |
| 11 |
Hb_158869_010 |
0.3374726851 |
- |
- |
PREDICTED: uncharacterized protein LOC105778995 [Gossypium raimondii] |
| 12 |
Hb_004584_010 |
0.3403484235 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 13 |
Hb_000608_130 |
0.3428022392 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_035905_010 |
0.3447562327 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 15 |
Hb_001495_060 |
0.3469394311 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 16 |
Hb_010812_090 |
0.3497458755 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 17 |
Hb_003335_070 |
0.3525575612 |
- |
- |
- |
| 18 |
Hb_005321_030 |
0.3536186034 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4 [Jatropha curcas] |
| 19 |
Hb_010883_100 |
0.3569151884 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 20 |
Hb_006502_040 |
0.3669347783 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |