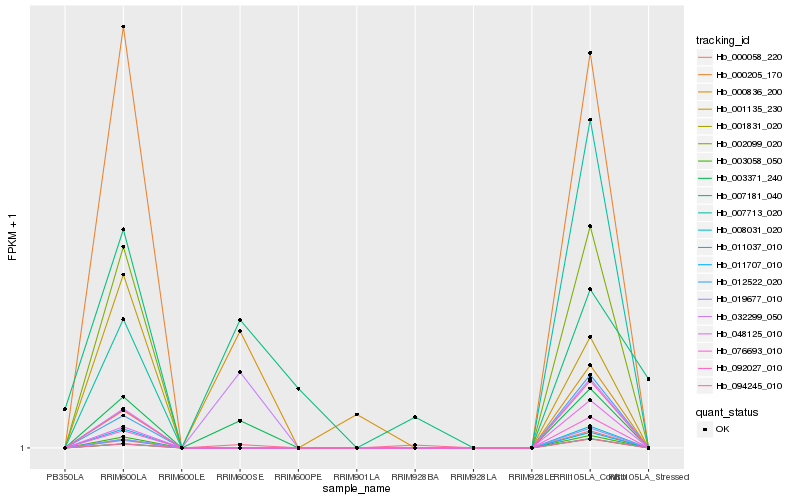
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_16702 [Jatropha curcas] |
| 2 |
Hb_032299_050 |
0.2211682496 |
- |
- |
- |
| 3 |
Hb_094245_010 |
0.3029493792 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 4 |
Hb_011037_010 |
0.3223357783 |
- |
- |
PREDICTED: uncharacterized protein LOC105778976 [Gossypium raimondii] |
| 5 |
Hb_003058_050 |
0.3223539275 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_002099_020 |
0.3223887701 |
- |
- |
cysteine proteinase inhibitor-like [Jatropha curcas] |
| 7 |
Hb_000205_170 |
0.3263133112 |
- |
- |
PREDICTED: uncharacterized protein LOC103926801 [Pyrus x bretschneideri] |
| 8 |
Hb_092027_010 |
0.3339157787 |
- |
- |
PREDICTED: uncharacterized protein LOC105761643 [Gossypium raimondii] |
| 9 |
Hb_076693_010 |
0.3340666576 |
- |
- |
PREDICTED: beta-glucosidase 11-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_011707_010 |
0.3341147904 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_008031_020 |
0.3467418874 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 12 |
Hb_001831_020 |
0.3469197589 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 13 |
Hb_000058_220 |
0.3472568374 |
- |
- |
hypothetical protein, partial [Staphylococcus aureus] |
| 14 |
Hb_019677_010 |
0.3473706639 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_048125_010 |
0.3485801341 |
- |
- |
PREDICTED: uncharacterized protein LOC104104928 [Nicotiana tomentosiformis] |
| 16 |
Hb_001135_230 |
0.3489763571 |
- |
- |
PREDICTED: protein REVEILLE 6-like isoform X3 [Gossypium raimondii] |
| 17 |
Hb_012522_020 |
0.3495693086 |
- |
- |
- |
| 18 |
Hb_007713_020 |
0.3740324299 |
- |
- |
- |
| 19 |
Hb_000836_200 |
0.383927999 |
- |
- |
- |
| 20 |
Hb_007181_040 |
0.3890711665 |
- |
- |
- |