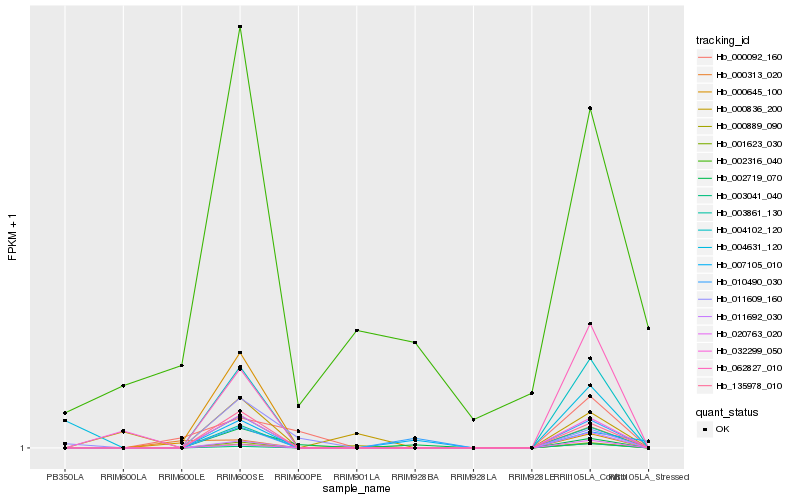
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003861_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_001623_030 |
0.0374942583 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_004102_120 |
0.044879665 |
- |
- |
- |
| 4 |
Hb_135978_010 |
0.0715701096 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 5 |
Hb_062827_010 |
0.1395508614 |
- |
- |
- |
| 6 |
Hb_011692_030 |
0.1885082736 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 7 |
Hb_020763_020 |
0.1895161792 |
- |
- |
hypothetical protein POPTR_0001s27010g [Populus trichocarpa] |
| 8 |
Hb_003041_040 |
0.2245189449 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_010490_030 |
0.302064756 |
- |
- |
- |
| 10 |
Hb_002719_070 |
0.3314838203 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 11 |
Hb_032299_050 |
0.3346314673 |
- |
- |
- |
| 12 |
Hb_000889_090 |
0.3383827676 |
- |
- |
isocitrate lyase, putative [Ricinus communis] |
| 13 |
Hb_011609_160 |
0.3446345137 |
- |
- |
Lipase, putative [Ricinus communis] |
| 14 |
Hb_007105_010 |
0.3729756694 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 15 |
Hb_000836_200 |
0.3743428447 |
- |
- |
- |
| 16 |
Hb_004631_120 |
0.3757952763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000645_100 |
0.3760015054 |
- |
- |
enzyme inhibitor, putative [Ricinus communis] |
| 18 |
Hb_000092_160 |
0.382075524 |
- |
- |
PREDICTED: 18 kDa seed maturation protein [Populus euphratica] |
| 19 |
Hb_000313_020 |
0.3856200707 |
- |
- |
Endoplasmic reticulum vesicle transporter protein isoform 2, partial [Theobroma cacao] |
| 20 |
Hb_002316_040 |
0.3866337517 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |