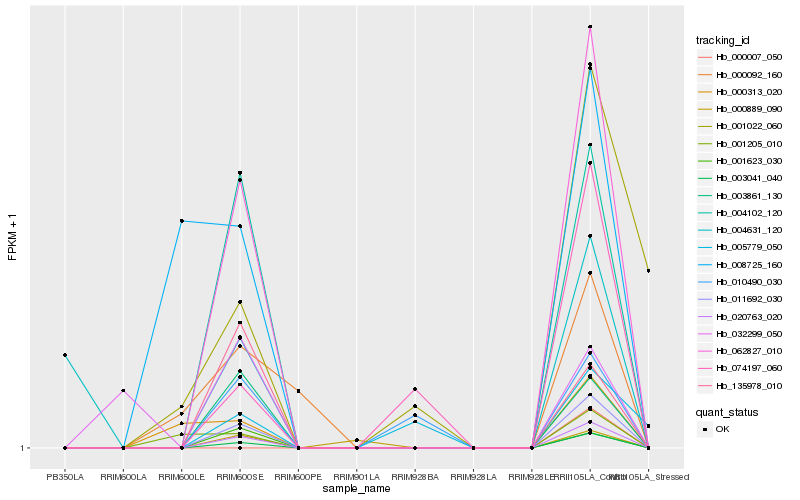
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011692_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 2 |
Hb_020763_020 |
0.0010222482 |
- |
- |
hypothetical protein POPTR_0001s27010g [Populus trichocarpa] |
| 3 |
Hb_003041_040 |
0.0366372585 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_062827_010 |
0.0494503724 |
- |
- |
- |
| 5 |
Hb_004102_120 |
0.1440673942 |
- |
- |
- |
| 6 |
Hb_003861_130 |
0.1885082736 |
- |
- |
- |
| 7 |
Hb_001623_030 |
0.2254439942 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_135978_010 |
0.258861619 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 9 |
Hb_074197_060 |
0.2960880555 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 isoform X1 [Populus euphratica] |
| 10 |
Hb_010490_030 |
0.3076090925 |
- |
- |
- |
| 11 |
Hb_000313_020 |
0.3247857414 |
- |
- |
Endoplasmic reticulum vesicle transporter protein isoform 2, partial [Theobroma cacao] |
| 12 |
Hb_001205_010 |
0.3325849017 |
- |
- |
- |
| 13 |
Hb_004631_120 |
0.3400902564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000889_090 |
0.3421441955 |
- |
- |
isocitrate lyase, putative [Ricinus communis] |
| 15 |
Hb_000092_160 |
0.360183393 |
- |
- |
PREDICTED: 18 kDa seed maturation protein [Populus euphratica] |
| 16 |
Hb_032299_050 |
0.3793663867 |
- |
- |
- |
| 17 |
Hb_008725_160 |
0.3799105927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005779_050 |
0.4037651675 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 19 |
Hb_001022_060 |
0.4137621272 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_000007_050 |
0.4155114937 |
- |
- |
- |