| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_200 |
0.0 |
- |
- |
- |
| 2 |
Hb_032299_050 |
0.2603370018 |
- |
- |
- |
| 3 |
Hb_000889_090 |
0.3017915653 |
- |
- |
isocitrate lyase, putative [Ricinus communis] |
| 4 |
Hb_004732_010 |
0.3220245404 |
- |
- |
- |
| 5 |
Hb_001059_020 |
0.3303524878 |
- |
- |
PREDICTED: FBD-associated F-box protein At4g10400-like [Malus domestica] |
| 6 |
Hb_002534_110 |
0.3462031501 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_006452_130 |
0.3469560072 |
- |
- |
- |
| 8 |
Hb_047287_040 |
0.349783258 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 9 |
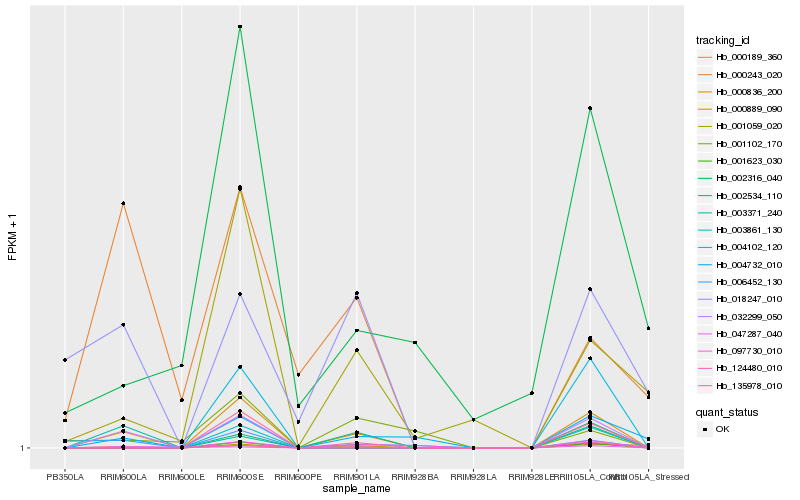
Hb_002316_040 |
0.3644404401 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 10 |
Hb_135978_010 |
0.3701517003 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 11 |
Hb_001623_030 |
0.3706791534 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_097730_010 |
0.3716303966 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 13 |
Hb_018247_010 |
0.3736669514 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 14 |
Hb_003861_130 |
0.3743428447 |
- |
- |
- |
| 15 |
Hb_000243_020 |
0.3814631722 |
- |
- |
- |
| 16 |
Hb_004102_120 |
0.3828228984 |
- |
- |
- |
| 17 |
Hb_000189_360 |
0.3834496981 |
- |
- |
Calcium-transporting ATPase 12, plasma membrane-type [Morus notabilis] |
| 18 |
Hb_003371_240 |
0.383927999 |
- |
- |
hypothetical protein JCGZ_16702 [Jatropha curcas] |
| 19 |
Hb_001102_170 |
0.3937753563 |
- |
- |
PREDICTED: protein FLX-like 1 [Jatropha curcas] |
| 20 |
Hb_124480_010 |
0.4062468796 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |