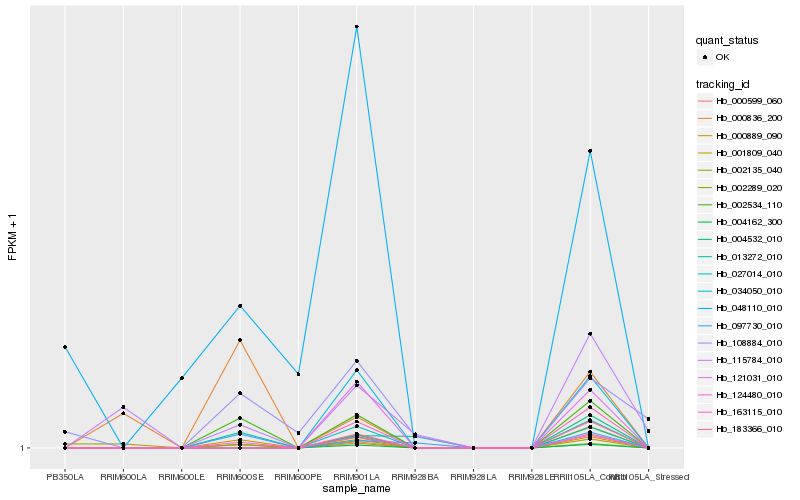
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_110 |
0.0 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_124480_010 |
0.091179032 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 3 |
Hb_000889_090 |
0.1024634682 |
- |
- |
isocitrate lyase, putative [Ricinus communis] |
| 4 |
Hb_002289_020 |
0.1929939245 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 5 |
Hb_097730_010 |
0.2756594766 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 6 |
Hb_013272_010 |
0.3236747855 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000836_200 |
0.3462031501 |
- |
- |
- |
| 8 |
Hb_048110_010 |
0.3690459151 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 9 |
Hb_108884_010 |
0.3789993034 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial [Prunus mume] |
| 10 |
Hb_163115_010 |
0.38651414 |
- |
- |
- |
| 11 |
Hb_027014_010 |
0.3866701495 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 12 |
Hb_004162_300 |
0.3871108451 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 13 |
Hb_183366_010 |
0.3901035329 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 14 |
Hb_001809_040 |
0.3920565207 |
- |
- |
PREDICTED: uncharacterized protein LOC105638660 [Jatropha curcas] |
| 15 |
Hb_000599_060 |
0.3950809552 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 16 |
Hb_034050_010 |
0.3961874269 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 17 |
Hb_121031_010 |
0.3984710778 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 18 |
Hb_004532_010 |
0.4012322183 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |
| 19 |
Hb_002135_040 |
0.4071617509 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 20 |
Hb_115784_010 |
0.4084319245 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |