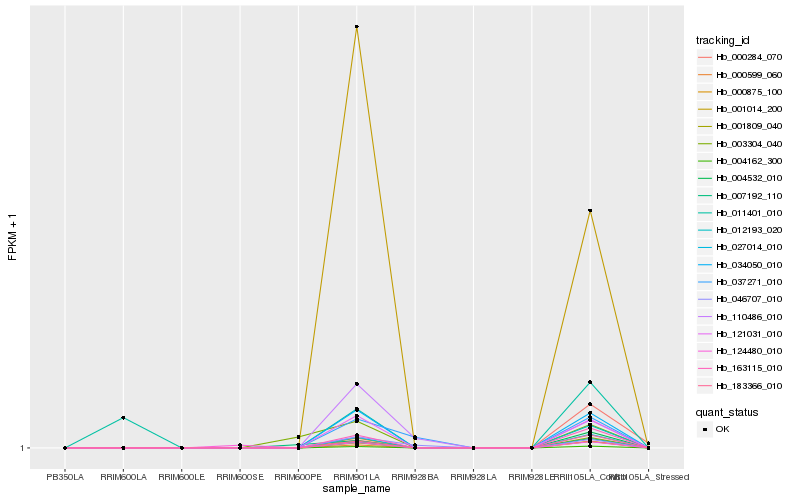
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_300 |
0.0 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 2 |
Hb_027014_010 |
0.0078867335 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_163115_010 |
0.0475932455 |
- |
- |
- |
| 4 |
Hb_034050_010 |
0.066915397 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 5 |
Hb_121031_010 |
0.0774965236 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 6 |
Hb_183366_010 |
0.0896313195 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 7 |
Hb_001809_040 |
0.103069283 |
- |
- |
PREDICTED: uncharacterized protein LOC105638660 [Jatropha curcas] |
| 8 |
Hb_000599_060 |
0.1200927066 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 9 |
Hb_000875_100 |
0.1384434618 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 10 |
Hb_004532_010 |
0.1475636935 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |
| 11 |
Hb_012193_020 |
0.22441951 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 12 |
Hb_046707_010 |
0.2703667882 |
- |
- |
PREDICTED: uncharacterized protein LOC104887079 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_001014_200 |
0.2719359912 |
- |
- |
- |
| 14 |
Hb_037271_010 |
0.2792426437 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 15 |
Hb_007192_110 |
0.2926313255 |
- |
- |
- |
| 16 |
Hb_110486_010 |
0.3112949218 |
- |
- |
- |
| 17 |
Hb_003304_040 |
0.3135547494 |
- |
- |
hypothetical protein CICLE_v10003343mg, partial [Citrus clementina] |
| 18 |
Hb_124480_010 |
0.3192478758 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 19 |
Hb_000284_070 |
0.346018198 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 20 |
Hb_011401_010 |
0.3479029637 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |