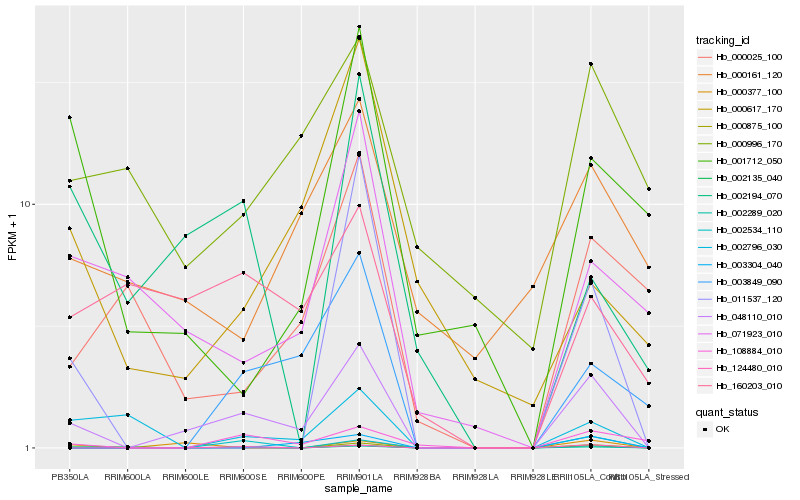
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048110_010 |
0.0 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 2 |
Hb_002289_020 |
0.3166419089 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 3 |
Hb_124480_010 |
0.3312192039 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_108884_010 |
0.3339425982 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial [Prunus mume] |
| 5 |
Hb_002135_040 |
0.3484231612 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 6 |
Hb_003849_090 |
0.3549749146 |
- |
- |
- |
| 7 |
Hb_071923_010 |
0.3588999915 |
- |
- |
- |
| 8 |
Hb_011537_120 |
0.3649545388 |
- |
- |
- |
| 9 |
Hb_160203_010 |
0.3670847472 |
- |
- |
- |
| 10 |
Hb_002534_110 |
0.3690459151 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000996_170 |
0.3698776738 |
- |
- |
unknown [Glycine max] |
| 12 |
Hb_002194_070 |
0.3706416167 |
- |
- |
- |
| 13 |
Hb_000377_100 |
0.3741871185 |
- |
- |
LINE-type retrotransposon LIb DNA, Insertion at the S11 site-like protein [Theobroma cacao] |
| 14 |
Hb_003304_040 |
0.3821455868 |
- |
- |
hypothetical protein CICLE_v10003343mg, partial [Citrus clementina] |
| 15 |
Hb_000161_120 |
0.3832446265 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000025_100 |
0.3852377966 |
- |
- |
- |
| 17 |
Hb_002796_030 |
0.3897326026 |
- |
- |
- |
| 18 |
Hb_000617_170 |
0.3898413683 |
- |
- |
- |
| 19 |
Hb_001712_050 |
0.3909346679 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 20 |
Hb_000875_100 |
0.3936922136 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |