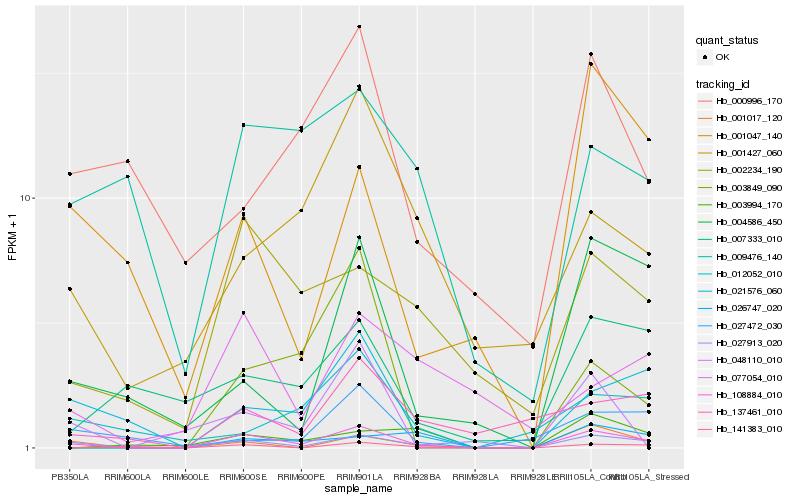
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108884_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial [Prunus mume] |
| 2 |
Hb_001017_120 |
0.2563194384 |
- |
- |
- |
| 3 |
Hb_027913_020 |
0.269065525 |
- |
- |
- |
| 4 |
Hb_021576_060 |
0.2799611365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002234_190 |
0.2941557248 |
- |
- |
PREDICTED: putative non-specific lipid-transfer protein 14 [Jatropha curcas] |
| 6 |
Hb_004586_450 |
0.3136992419 |
- |
- |
- |
| 7 |
Hb_137461_010 |
0.3200045801 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 8 |
Hb_001427_060 |
0.3205778495 |
- |
- |
- |
| 9 |
Hb_141383_010 |
0.3246216845 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 10 |
Hb_009476_140 |
0.3299507833 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_0838610 [Ricinus communis] |
| 11 |
Hb_048110_010 |
0.3339425982 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 12 |
Hb_026747_020 |
0.3355670654 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_007333_010 |
0.3356023645 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_012052_010 |
0.3365412795 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 15 |
Hb_027472_030 |
0.336599749 |
- |
- |
RNA helicase 36, partial [Manihot esculenta] |
| 16 |
Hb_000996_170 |
0.3375344289 |
- |
- |
unknown [Glycine max] |
| 17 |
Hb_001047_140 |
0.3384451821 |
- |
- |
- |
| 18 |
Hb_003994_170 |
0.3411236808 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 19 |
Hb_077054_010 |
0.3411965057 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 20 |
Hb_003849_090 |
0.3425474725 |
- |
- |
- |