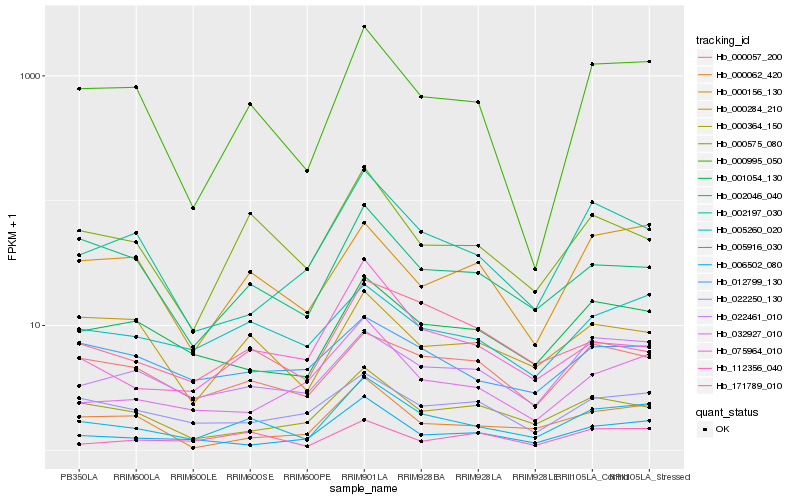
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005916_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 2 |
Hb_000995_050 |
0.1337974549 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 3 |
Hb_000575_080 |
0.1340548566 |
- |
- |
PREDICTED: CD2 antigen cytoplasmic tail-binding protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005260_020 |
0.1438719937 |
- |
- |
PREDICTED: uncharacterized protein LOC105633789 [Jatropha curcas] |
| 5 |
Hb_022461_010 |
0.1558174021 |
- |
- |
- |
| 6 |
Hb_001054_130 |
0.1558696961 |
- |
- |
Protein SSU72, putative [Ricinus communis] |
| 7 |
Hb_006502_080 |
0.1597536278 |
- |
- |
- |
| 8 |
Hb_002046_040 |
0.1616602024 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |
| 9 |
Hb_000057_200 |
0.1645063735 |
- |
- |
PREDICTED: methyltransferase-like protein 22 [Jatropha curcas] |
| 10 |
Hb_032927_010 |
0.1653284185 |
- |
- |
- |
| 11 |
Hb_075964_010 |
0.1653389491 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 12 |
Hb_000062_420 |
0.1658494746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000364_150 |
0.1682695858 |
- |
- |
- |
| 14 |
Hb_000156_130 |
0.169578799 |
- |
- |
hypothetical protein JCGZ_04605 [Jatropha curcas] |
| 15 |
Hb_112356_040 |
0.1721200927 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 16 |
Hb_012799_130 |
0.1722817288 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 17 |
Hb_022250_130 |
0.1728589199 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |
| 18 |
Hb_000284_210 |
0.1743060114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_171789_010 |
0.1750131889 |
- |
- |
phosphoenolpyruvate carboxylase [Glycine max] |
| 20 |
Hb_002197_030 |
0.1753835233 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X2 [Jatropha curcas] |