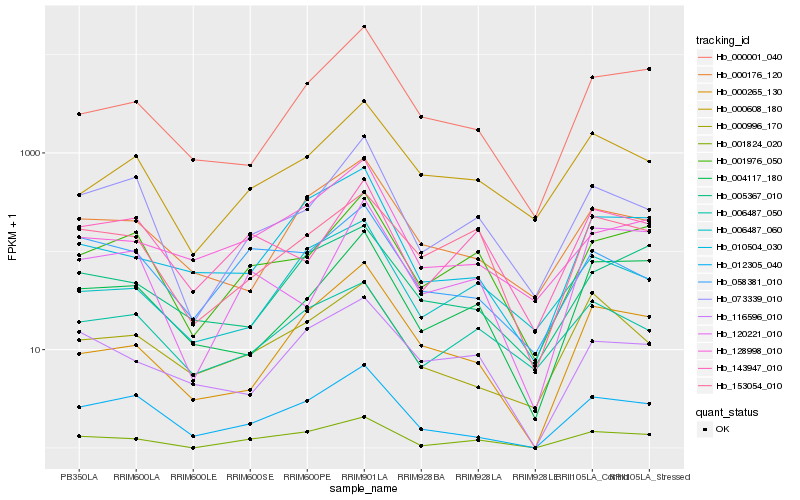
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001824_020 |
0.0 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 2 |
Hb_001976_050 |
0.172876199 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 3 |
Hb_012305_040 |
0.1773577482 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A-like [Jatropha curcas] |
| 4 |
Hb_153054_010 |
0.1775705975 |
- |
- |
PREDICTED: metal tolerance protein 11-like [Populus euphratica] |
| 5 |
Hb_073339_010 |
0.1860074396 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 6 |
Hb_006487_060 |
0.1872373251 |
- |
- |
Defective in cullin neddylation protein, putative [Ricinus communis] |
| 7 |
Hb_010504_030 |
0.1897235619 |
- |
- |
calmodulin homolog {EST} [Brassica napus, Naehan, root, Peptide Partial, 53 aa] |
| 8 |
Hb_006487_050 |
0.1973567335 |
- |
- |
hypothetical protein PHAVU_005G049600g [Phaseolus vulgaris] |
| 9 |
Hb_116596_010 |
0.199199845 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070-like isoform X1 [Citrus sinensis] |
| 10 |
Hb_004117_180 |
0.2002517021 |
- |
- |
- |
| 11 |
Hb_143947_010 |
0.2009993231 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X2 [Pyrus x bretschneideri] |
| 12 |
Hb_000265_130 |
0.2013944453 |
- |
- |
- |
| 13 |
Hb_058381_010 |
0.2036668977 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 14 |
Hb_120221_010 |
0.2037746036 |
- |
- |
- |
| 15 |
Hb_000608_180 |
0.2058029798 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |
| 16 |
Hb_000001_040 |
0.2098816123 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000176_120 |
0.2108175773 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |
| 18 |
Hb_128998_010 |
0.2111119849 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 19 |
Hb_005367_010 |
0.21118041 |
- |
- |
- |
| 20 |
Hb_000996_170 |
0.211384235 |
- |
- |
unknown [Glycine max] |