| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
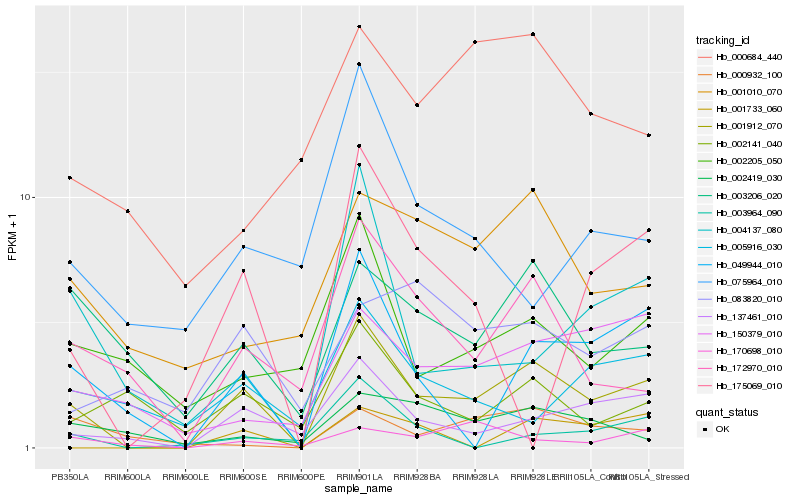
Hb_001912_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s03380g [Populus trichocarpa] |
| 2 |
Hb_137461_010 |
0.2385081479 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 3 |
Hb_049944_010 |
0.2566367909 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 4 |
Hb_172970_010 |
0.2680143133 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 5 |
Hb_003206_020 |
0.2765840065 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 6 |
Hb_004137_080 |
0.2834255348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_150379_010 |
0.2851943093 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 8 |
Hb_002205_050 |
0.2893863626 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 9 |
Hb_001010_070 |
0.2912715965 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 10 |
Hb_002141_040 |
0.3038251737 |
- |
- |
- |
| 11 |
Hb_003964_090 |
0.3065131248 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 12 |
Hb_001733_060 |
0.3075596832 |
- |
- |
PREDICTED: uncharacterized protein LOC104431099 [Eucalyptus grandis] |
| 13 |
Hb_005916_030 |
0.3076086419 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 14 |
Hb_002419_030 |
0.3079444986 |
- |
- |
- |
| 15 |
Hb_170698_010 |
0.3103867434 |
- |
- |
PREDICTED: uncharacterized protein LOC104120414 [Nicotiana tomentosiformis] |
| 16 |
Hb_075964_010 |
0.312436126 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 17 |
Hb_000684_440 |
0.3136851958 |
- |
- |
hypothetical protein CICLE_v100111082mg, partial [Citrus clementina] |
| 18 |
Hb_000932_100 |
0.314672883 |
- |
- |
- |
| 19 |
Hb_083820_010 |
0.3149758785 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 20 |
Hb_175069_010 |
0.3199974542 |
- |
- |
hypothetical protein POPTR_0002s210401g, partial [Populus trichocarpa] |