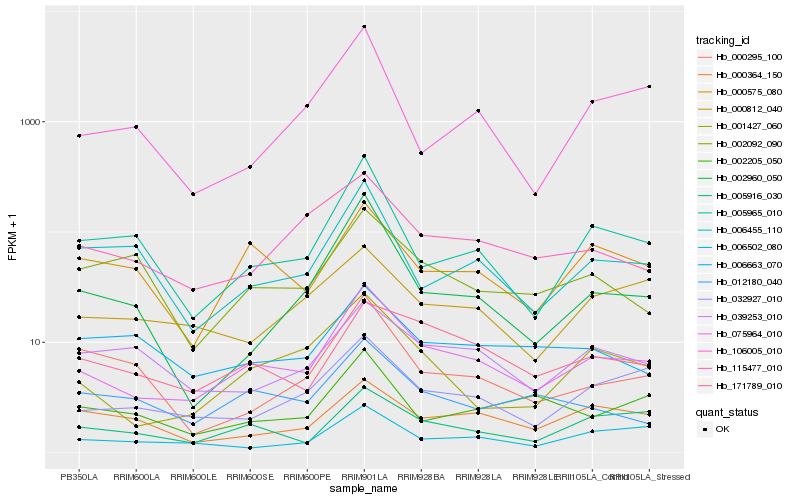
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075964_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_001427_060 |
0.1580933952 |
- |
- |
- |
| 3 |
Hb_171789_010 |
0.1605890537 |
- |
- |
phosphoenolpyruvate carboxylase [Glycine max] |
| 4 |
Hb_032927_010 |
0.1619052052 |
- |
- |
- |
| 5 |
Hb_005916_030 |
0.1653389491 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 6 |
Hb_005965_010 |
0.1705667241 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 7 |
Hb_115477_010 |
0.1711261666 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 8 |
Hb_006502_080 |
0.1778561906 |
- |
- |
- |
| 9 |
Hb_000575_080 |
0.1786715737 |
- |
- |
PREDICTED: CD2 antigen cytoplasmic tail-binding protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002960_050 |
0.1799323106 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 11 |
Hb_012180_040 |
0.1822920716 |
- |
- |
hypothetical protein VITISV_042517 [Vitis vinifera] |
| 12 |
Hb_039253_010 |
0.184524917 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006455_110 |
0.1849870346 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000812_040 |
0.1883690527 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 15 |
Hb_000364_150 |
0.1953904161 |
- |
- |
- |
| 16 |
Hb_006663_070 |
0.196215843 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 17 |
Hb_002205_050 |
0.1963930291 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 18 |
Hb_002092_090 |
0.1978686261 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 19 |
Hb_000295_100 |
0.198335414 |
- |
- |
- |
| 20 |
Hb_106005_010 |
0.199495062 |
- |
- |
EG5651 [Manihot esculenta] |