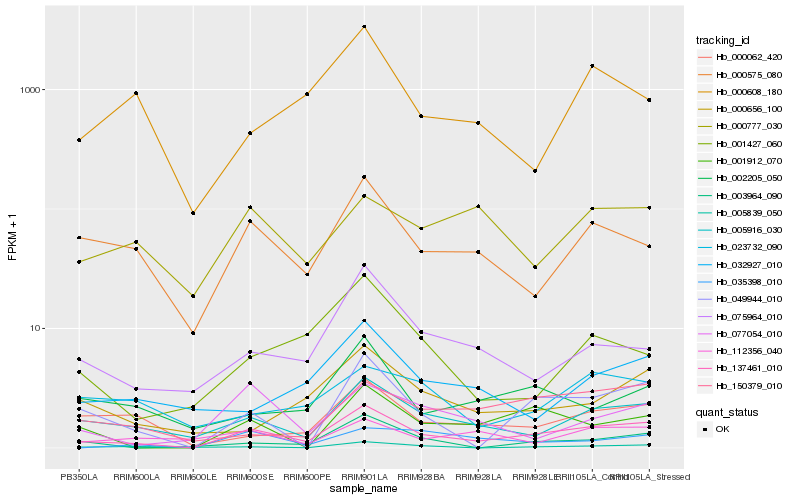
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137461_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 2 |
Hb_005916_030 |
0.1919627688 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 3 |
Hb_049944_010 |
0.2200119725 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 4 |
Hb_000575_080 |
0.2285321718 |
- |
- |
PREDICTED: CD2 antigen cytoplasmic tail-binding protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_075964_010 |
0.2342209753 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 6 |
Hb_000062_420 |
0.2356820909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001912_070 |
0.2385081479 |
- |
- |
hypothetical protein POPTR_0003s03380g [Populus trichocarpa] |
| 8 |
Hb_150379_010 |
0.2412857876 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 9 |
Hb_000656_100 |
0.247240131 |
- |
- |
PREDICTED: uncharacterized protein LOC105633651 [Jatropha curcas] |
| 10 |
Hb_003964_090 |
0.2475211051 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 11 |
Hb_002205_050 |
0.247998005 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 12 |
Hb_005839_050 |
0.2483931456 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 13 |
Hb_001427_060 |
0.2500183932 |
- |
- |
- |
| 14 |
Hb_032927_010 |
0.2520402649 |
- |
- |
- |
| 15 |
Hb_077054_010 |
0.2531447986 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 16 |
Hb_112356_040 |
0.2603314933 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 17 |
Hb_035398_010 |
0.2604626136 |
- |
- |
hypothetical protein JCGZ_13029 [Jatropha curcas] |
| 18 |
Hb_023732_090 |
0.2620837392 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000777_030 |
0.2629075137 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 20 |
Hb_000608_180 |
0.2629966185 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |