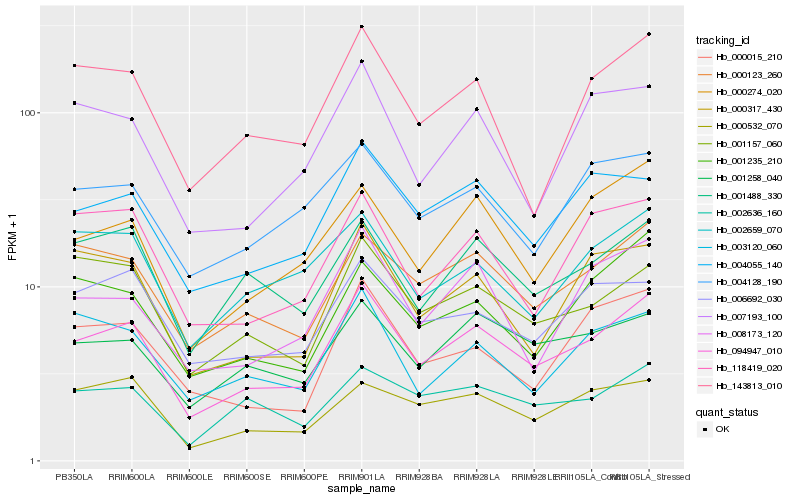
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094947_010 |
0.0 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 2 |
Hb_000123_260 |
0.0969301318 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 3 |
Hb_004055_140 |
0.1007058627 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 4 |
Hb_001258_040 |
0.1059638101 |
- |
- |
hypothetical protein JCGZ_12578 [Jatropha curcas] |
| 5 |
Hb_001488_330 |
0.1085282316 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 6 |
Hb_000274_020 |
0.1088173702 |
- |
- |
PREDICTED: uncharacterized protein LOC105630805 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001157_060 |
0.1088661083 |
- |
- |
PREDICTED: putative nitric oxide synthase [Jatropha curcas] |
| 8 |
Hb_143813_010 |
0.1094238931 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1 [Nelumbo nucifera] |
| 9 |
Hb_008173_120 |
0.110604831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004128_190 |
0.1148907217 |
- |
- |
PREDICTED: dirigent protein 17-like [Jatropha curcas] |
| 11 |
Hb_000317_430 |
0.114899082 |
transcription factor |
TF Family: SET |
PREDICTED: ribosomal lysine N-methyltransferase 3 [Jatropha curcas] |
| 12 |
Hb_006692_030 |
0.1166043728 |
- |
- |
PREDICTED: uncharacterized protein LOC105632913 [Jatropha curcas] |
| 13 |
Hb_002636_160 |
0.116719154 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g13630 [Jatropha curcas] |
| 14 |
Hb_118419_020 |
0.1169931006 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Vitis vinifera] |
| 15 |
Hb_000015_210 |
0.1185876162 |
- |
- |
PREDICTED: uncharacterized protein LOC105647781 [Jatropha curcas] |
| 16 |
Hb_000532_070 |
0.1186375282 |
- |
- |
PREDICTED: exosome complex component RRP42-like [Jatropha curcas] |
| 17 |
Hb_003120_060 |
0.1187258501 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 18 |
Hb_007193_100 |
0.1192191375 |
- |
- |
syntaxin family protein [Populus trichocarpa] |
| 19 |
Hb_002659_070 |
0.1194771189 |
- |
- |
PREDICTED: RNA-binding protein 24-A [Jatropha curcas] |
| 20 |
Hb_001235_210 |
0.1195587212 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |