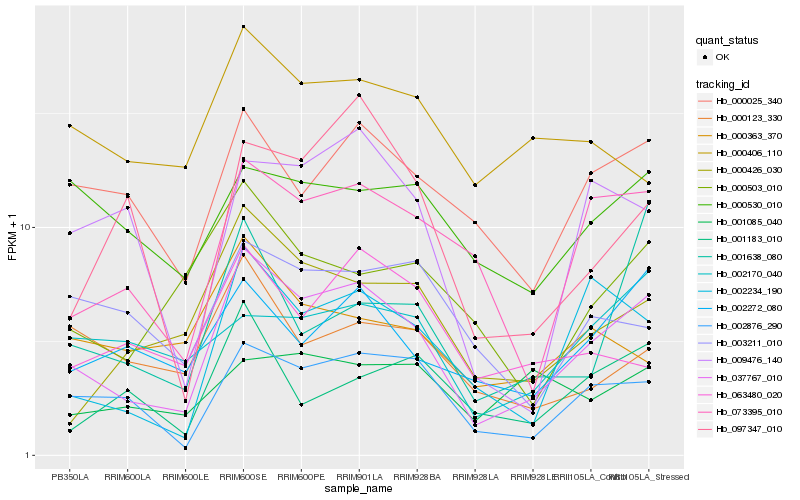
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_037767_010 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_002876_290 |
0.1584672457 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 3 |
Hb_002234_190 |
0.1742279656 |
- |
- |
PREDICTED: putative non-specific lipid-transfer protein 14 [Jatropha curcas] |
| 4 |
Hb_073395_010 |
0.1745432662 |
- |
- |
- |
| 5 |
Hb_000025_340 |
0.1837908337 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 6 |
Hb_000426_030 |
0.1845838792 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002272_080 |
0.189865479 |
- |
- |
- |
| 8 |
Hb_000503_010 |
0.1979729208 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 9 |
Hb_009476_140 |
0.2086945519 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_0838610 [Ricinus communis] |
| 10 |
Hb_000123_330 |
0.2093317397 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 11 |
Hb_002170_040 |
0.2145224112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000530_010 |
0.2152805188 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 13 |
Hb_001183_010 |
0.2154141118 |
- |
- |
- |
| 14 |
Hb_063480_020 |
0.2155267306 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000363_370 |
0.2156933747 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 16 |
Hb_003211_010 |
0.2167856987 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 17 |
Hb_097347_010 |
0.2197294697 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like, partial [Malus domestica] |
| 18 |
Hb_000406_110 |
0.2200834956 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 19 |
Hb_001638_080 |
0.2204029501 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 20 |
Hb_001085_040 |
0.2231017169 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |