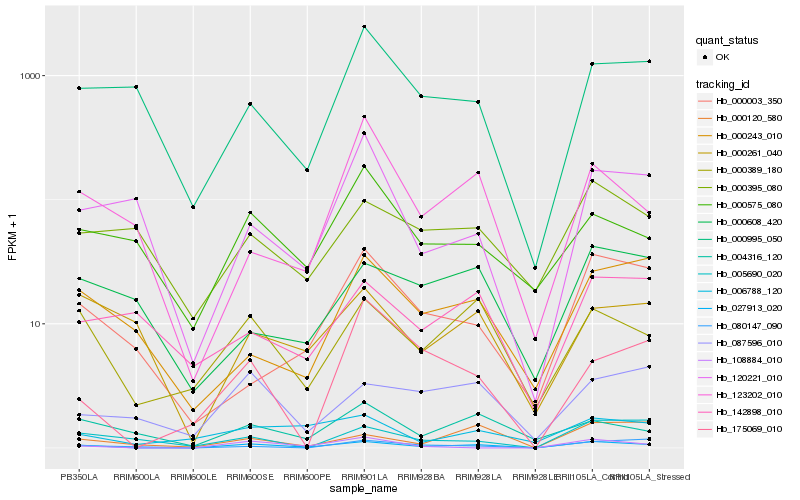
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027913_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000389_180 |
0.215916188 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_000120_580 |
0.259714508 |
- |
- |
hypothetical protein POPTR_0007s08570g [Populus trichocarpa] |
| 4 |
Hb_087596_010 |
0.2679673818 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 5 |
Hb_108884_010 |
0.269065525 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial [Prunus mume] |
| 6 |
Hb_004316_120 |
0.2696969198 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Populus euphratica] |
| 7 |
Hb_006788_120 |
0.2758138697 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000995_050 |
0.2802224174 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 9 |
Hb_000243_010 |
0.2803754596 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 10 |
Hb_000003_350 |
0.2812128934 |
- |
- |
hypothetical protein POPTR_1155s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_000261_040 |
0.2840296725 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 12 |
Hb_005690_020 |
0.286060986 |
- |
- |
hypothetical protein POPTR_0006s08200g [Populus trichocarpa] |
| 13 |
Hb_123202_010 |
0.2865185716 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 14 |
Hb_000575_080 |
0.2875281115 |
- |
- |
PREDICTED: CD2 antigen cytoplasmic tail-binding protein 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_080147_090 |
0.2893581282 |
- |
- |
hypothetical protein JCGZ_07271 [Jatropha curcas] |
| 16 |
Hb_000395_080 |
0.289367191 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 17 |
Hb_175069_010 |
0.2899936853 |
- |
- |
hypothetical protein POPTR_0002s210401g, partial [Populus trichocarpa] |
| 18 |
Hb_120221_010 |
0.2905793136 |
- |
- |
- |
| 19 |
Hb_000608_420 |
0.2921760627 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 20 |
Hb_142898_010 |
0.2933105967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |