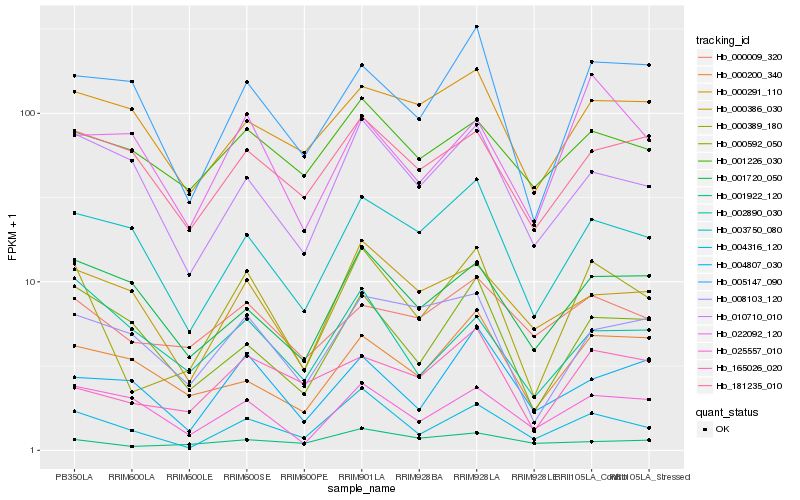
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000389_180 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_004316_120 |
0.1578625172 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Populus euphratica] |
| 3 |
Hb_165026_020 |
0.1793286742 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0014s10300g [Populus trichocarpa] |
| 4 |
Hb_003750_080 |
0.1861721546 |
- |
- |
PREDICTED: DNA repair protein complementing XP-C cells homolog [Jatropha curcas] |
| 5 |
Hb_005147_090 |
0.1922731603 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008103_120 |
0.1928017888 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 7 |
Hb_000592_050 |
0.1929218104 |
- |
- |
PREDICTED: golgin candidate 2 [Jatropha curcas] |
| 8 |
Hb_000009_320 |
0.1980348823 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66631 [Jatropha curcas] |
| 9 |
Hb_010710_010 |
0.2000978991 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 10 |
Hb_025557_010 |
0.2025329025 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein DOT4, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000200_340 |
0.2038164772 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 12 |
Hb_000386_030 |
0.2045106206 |
- |
- |
PREDICTED: uncharacterized protein LOC105629270 [Jatropha curcas] |
| 13 |
Hb_001720_050 |
0.2051038393 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001922_120 |
0.2062125979 |
- |
- |
hypothetical protein B456_004G136900 [Gossypium raimondii] |
| 15 |
Hb_181235_010 |
0.2074880926 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105645554 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002890_030 |
0.2075817742 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 17 |
Hb_001226_030 |
0.2083181852 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 [Jatropha curcas] |
| 18 |
Hb_022092_120 |
0.2093863371 |
- |
- |
PREDICTED: uncharacterized protein LOC105646971 [Jatropha curcas] |
| 19 |
Hb_004807_030 |
0.2096810913 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 20 |
Hb_000291_110 |
0.2107877212 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |