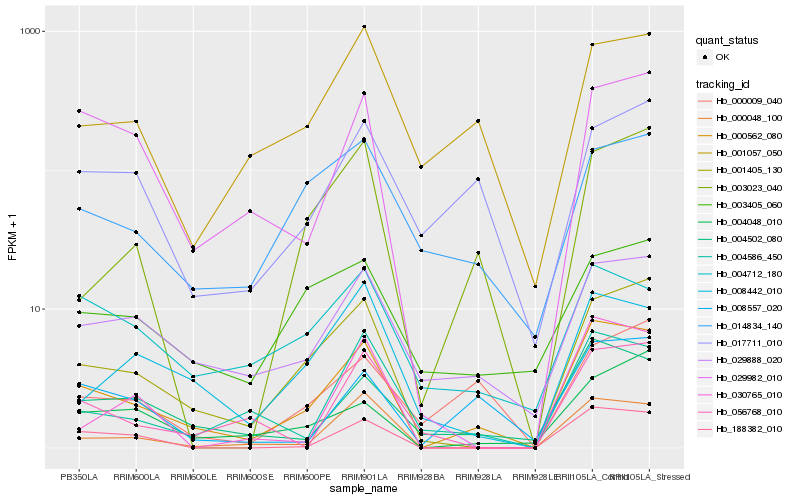
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000562_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001405_130 |
0.1520648361 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 3 |
Hb_004502_080 |
0.1789470453 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 4 |
Hb_029888_020 |
0.1818802587 |
- |
- |
hypothetical protein PHAVU_007G268100g [Phaseolus vulgaris] |
| 5 |
Hb_000048_100 |
0.18431499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_188382_010 |
0.1854544663 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 7 |
Hb_008557_020 |
0.1988474259 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 8 |
Hb_029982_010 |
0.2019101033 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 9 |
Hb_004586_450 |
0.2020026547 |
- |
- |
- |
| 10 |
Hb_004048_010 |
0.2090762089 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 11 |
Hb_008442_010 |
0.2101423547 |
- |
- |
PREDICTED: nudix hydrolase 16, mitochondrial-like [Citrus sinensis] |
| 12 |
Hb_003023_040 |
0.2149275401 |
- |
- |
- |
| 13 |
Hb_001057_050 |
0.2157449214 |
- |
- |
hypothetical protein POPTR_0003s15960g, partial [Populus trichocarpa] |
| 14 |
Hb_030765_010 |
0.2201475294 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 15 |
Hb_000009_040 |
0.220346609 |
- |
- |
- |
| 16 |
Hb_014834_140 |
0.2211796945 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 17 |
Hb_004712_180 |
0.2241883348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003405_060 |
0.2256196504 |
- |
- |
- |
| 19 |
Hb_017711_010 |
0.2263063591 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 20 |
Hb_056768_010 |
0.231848227 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Cicer arietinum] |