| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
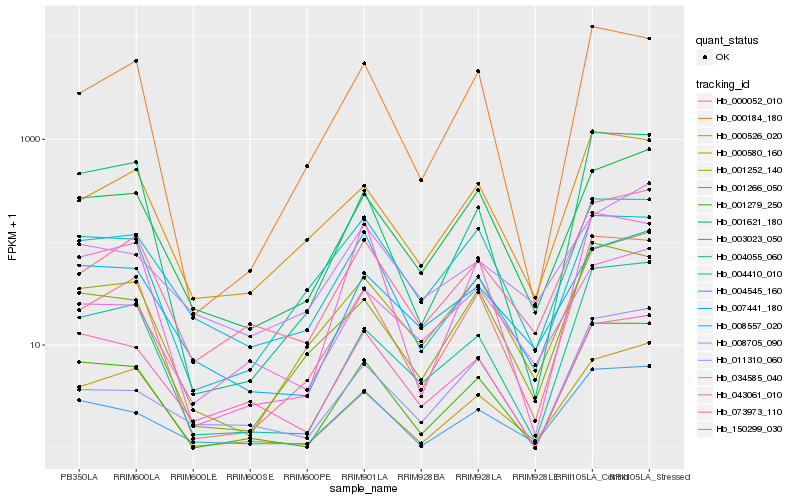
Hb_008557_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 2 |
Hb_001279_250 |
0.1176110259 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 3 |
Hb_001621_180 |
0.1455291447 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-1 [Jatropha curcas] |
| 4 |
Hb_001266_050 |
0.1479000045 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like isoform X2 [Elaeis guineensis] |
| 5 |
Hb_150299_030 |
0.1508483186 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001252_140 |
0.1511232099 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000052_010 |
0.1520252567 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 8 |
Hb_004410_010 |
0.1528164362 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 9 |
Hb_004545_160 |
0.158774981 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 10 |
Hb_007441_180 |
0.161792388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_043061_010 |
0.1619072237 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 12 |
Hb_073973_110 |
0.1650185295 |
- |
- |
PREDICTED: uncharacterized protein LOC105638912 [Jatropha curcas] |
| 13 |
Hb_011310_060 |
0.1655094659 |
- |
- |
PREDICTED: uncharacterized protein At2g34160-like [Jatropha curcas] |
| 14 |
Hb_000184_180 |
0.1661311438 |
- |
- |
putative C3HC4-type RING zinc finger protein [Hevea brasiliensis] |
| 15 |
Hb_004055_060 |
0.1663126907 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 16 |
Hb_003023_050 |
0.1665430097 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 17 |
Hb_008705_090 |
0.1667221085 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2-like [Jatropha curcas] |
| 18 |
Hb_000526_020 |
0.1667653929 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 19 |
Hb_034585_040 |
0.1672396283 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 20 |
Hb_000580_160 |
0.1679606525 |
- |
- |
Nitrate transporter 1.5 [Morus notabilis] |