| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
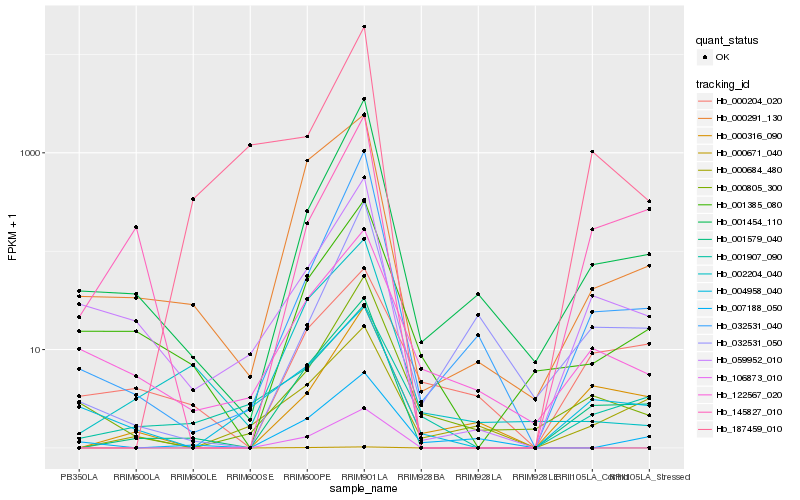
Hb_001579_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000291_130 |
0.163642343 |
- |
- |
- |
| 3 |
Hb_001454_110 |
0.1756872189 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 4 |
Hb_059952_010 |
0.1869017628 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 5 |
Hb_000316_090 |
0.1942184949 |
- |
- |
- |
| 6 |
Hb_032531_040 |
0.1946877346 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 7 |
Hb_145827_010 |
0.1995323286 |
- |
- |
hypothetical protein POPTR_0003s13630g [Populus trichocarpa] |
| 8 |
Hb_001385_080 |
0.2112010001 |
- |
- |
- |
| 9 |
Hb_001907_090 |
0.214637693 |
- |
- |
- |
| 10 |
Hb_002204_040 |
0.2149332638 |
- |
- |
hypothetical protein B456_008G181200 [Gossypium raimondii] |
| 11 |
Hb_000684_480 |
0.2151212465 |
transcription factor |
TF Family: Orphans |
- |
| 12 |
Hb_187459_010 |
0.2164521133 |
- |
- |
PREDICTED: uncharacterized protein LOC102619199, partial [Citrus sinensis] |
| 13 |
Hb_000805_300 |
0.2193897542 |
- |
- |
- |
| 14 |
Hb_106873_010 |
0.221125234 |
- |
- |
- |
| 15 |
Hb_122567_020 |
0.2226635961 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 16 |
Hb_004958_040 |
0.2285337174 |
- |
- |
- |
| 17 |
Hb_032531_050 |
0.2401417218 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 18 |
Hb_000671_040 |
0.2497239803 |
- |
- |
- |
| 19 |
Hb_000204_020 |
0.250502805 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 20 |
Hb_007188_050 |
0.251537322 |
- |
- |
- |