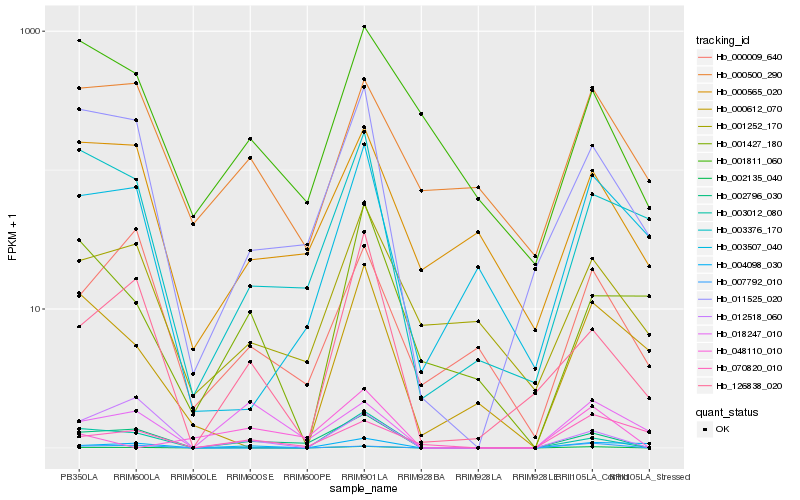
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002135_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 2 |
Hb_002796_030 |
0.2206422938 |
- |
- |
- |
| 3 |
Hb_018247_010 |
0.2729819699 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 4 |
Hb_004098_030 |
0.2982699651 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_126838_020 |
0.3009772708 |
- |
- |
JHL22C18.7 [Jatropha curcas] |
| 6 |
Hb_070820_010 |
0.3077794946 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 7 |
Hb_011525_020 |
0.3108503171 |
- |
- |
PREDICTED: uncharacterized protein LOC105640903 [Jatropha curcas] |
| 8 |
Hb_003012_080 |
0.3205145829 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_007792_010 |
0.3222648308 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 10 |
Hb_000500_290 |
0.333866685 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_012518_060 |
0.3341999901 |
- |
- |
- |
| 12 |
Hb_001252_170 |
0.3433223728 |
desease resistance |
Gene Name: Exostosin |
hypothetical protein PRUPE_ppa001357mg [Prunus persica] |
| 13 |
Hb_003376_170 |
0.3444867954 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000009_640 |
0.3452729411 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_12318 [Jatropha curcas] |
| 15 |
Hb_048110_010 |
0.3484231612 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 16 |
Hb_001811_060 |
0.3518508264 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003507_040 |
0.3527330552 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 18 |
Hb_001427_180 |
0.3552489514 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 19 |
Hb_000565_020 |
0.3569173222 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 20 |
Hb_000612_070 |
0.356943131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |