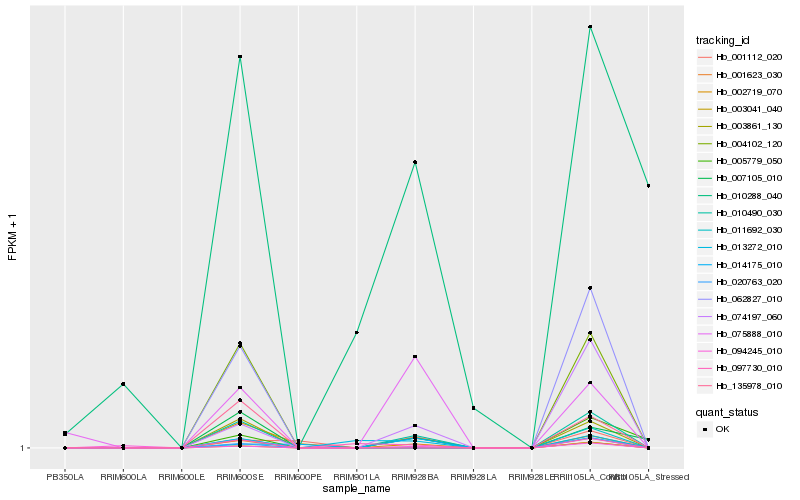
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010490_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_074197_060 |
0.2359825761 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 isoform X1 [Populus euphratica] |
| 3 |
Hb_005779_050 |
0.2833034232 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 4 |
Hb_007105_010 |
0.2897553016 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 5 |
Hb_004102_120 |
0.293483364 |
- |
- |
- |
| 6 |
Hb_062827_010 |
0.2956859324 |
- |
- |
- |
| 7 |
Hb_003861_130 |
0.302064756 |
- |
- |
- |
| 8 |
Hb_097730_010 |
0.3047052626 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 9 |
Hb_002719_070 |
0.3069295186 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 10 |
Hb_011692_030 |
0.3076090925 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 11 |
Hb_020763_020 |
0.3079266628 |
- |
- |
hypothetical protein POPTR_0001s27010g [Populus trichocarpa] |
| 12 |
Hb_075888_010 |
0.3122800718 |
- |
- |
PREDICTED: 25S rRNA (cytosine-C(5))-methyltransferase nop2-like [Solanum lycopersicum] |
| 13 |
Hb_001623_030 |
0.3135434579 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_003041_040 |
0.3206339655 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_135978_010 |
0.3270022136 |
- |
- |
hypothetical protein EUGRSUZ_J026621, partial [Eucalyptus grandis] |
| 16 |
Hb_001112_020 |
0.3390142676 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 17 |
Hb_013272_010 |
0.3418671712 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_010288_040 |
0.3561495442 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 19 |
Hb_094245_010 |
0.3577401838 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 20 |
Hb_014175_010 |
0.3698239327 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |