| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
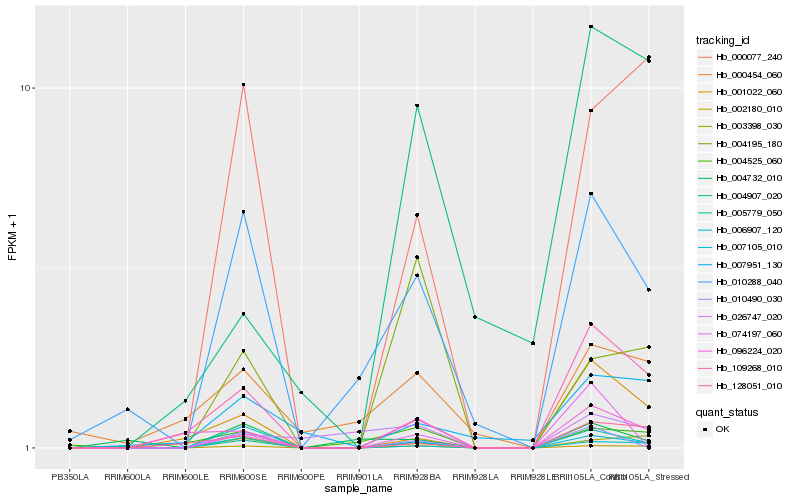
Hb_005779_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 2 |
Hb_109268_010 |
0.2012988555 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 3 |
Hb_001022_060 |
0.2337638193 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_010288_040 |
0.2455146909 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 5 |
Hb_007105_010 |
0.2621533242 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 6 |
Hb_006907_120 |
0.2709525812 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 7 |
Hb_128051_010 |
0.282487529 |
- |
- |
- |
| 8 |
Hb_096224_020 |
0.2828280453 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 9 |
Hb_000077_240 |
0.2830772778 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 10 |
Hb_010490_030 |
0.2833034232 |
- |
- |
- |
| 11 |
Hb_003398_030 |
0.2853170394 |
- |
- |
PREDICTED: uncharacterized protein LOC104583892 [Brachypodium distachyon] |
| 12 |
Hb_074197_060 |
0.2964378881 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 isoform X1 [Populus euphratica] |
| 13 |
Hb_002180_010 |
0.3098009053 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_007951_130 |
0.3396253873 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 15 |
Hb_004907_020 |
0.3486766219 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 16 |
Hb_004525_060 |
0.3488683875 |
- |
- |
- |
| 17 |
Hb_004732_010 |
0.3602223679 |
- |
- |
- |
| 18 |
Hb_000454_060 |
0.3667372199 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 19 |
Hb_004195_180 |
0.3688723817 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_026747_020 |
0.3724367123 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |