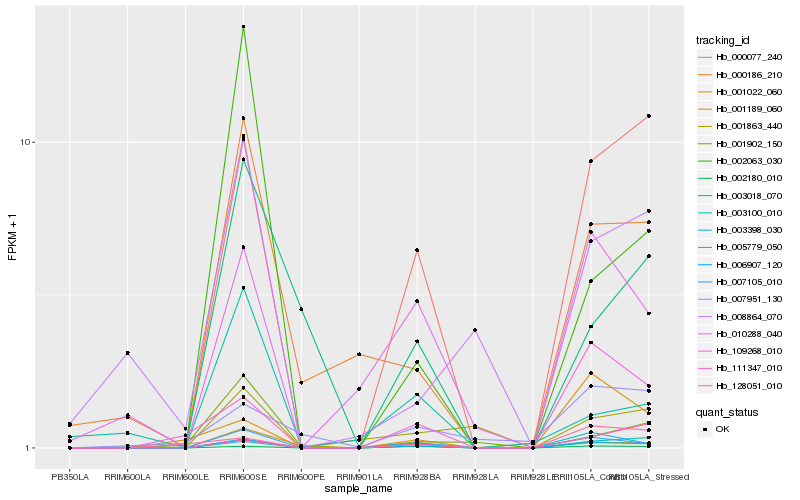
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006907_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 2 |
Hb_007105_010 |
0.1272707574 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 3 |
Hb_000077_240 |
0.163608797 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 4 |
Hb_003398_030 |
0.1848767425 |
- |
- |
PREDICTED: uncharacterized protein LOC104583892 [Brachypodium distachyon] |
| 5 |
Hb_001189_060 |
0.2338616211 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 6 |
Hb_000186_210 |
0.2431022477 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 7 |
Hb_010288_040 |
0.2679585524 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 8 |
Hb_005779_050 |
0.2709525812 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 9 |
Hb_002063_030 |
0.2833181362 |
- |
- |
- |
| 10 |
Hb_109268_010 |
0.2858941394 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 11 |
Hb_001902_150 |
0.2982104295 |
- |
- |
PREDICTED: cytokinin hydroxylase-like [Jatropha curcas] |
| 12 |
Hb_003018_070 |
0.2999403012 |
- |
- |
SAUR family protein, partial [Theobroma cacao] |
| 13 |
Hb_001863_440 |
0.3006074888 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 14 |
Hb_008864_070 |
0.3020227883 |
- |
- |
PREDICTED: uncharacterized protein LOC105803851 [Gossypium raimondii] |
| 15 |
Hb_128051_010 |
0.3060559203 |
- |
- |
- |
| 16 |
Hb_111347_010 |
0.3201393316 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 17 |
Hb_001022_060 |
0.325281873 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 18 |
Hb_003100_010 |
0.3305755345 |
- |
- |
PREDICTED: uncharacterized protein LOC101217345 [Cucumis sativus] |
| 19 |
Hb_007951_130 |
0.3308875004 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 20 |
Hb_002180_010 |
0.3318954646 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |