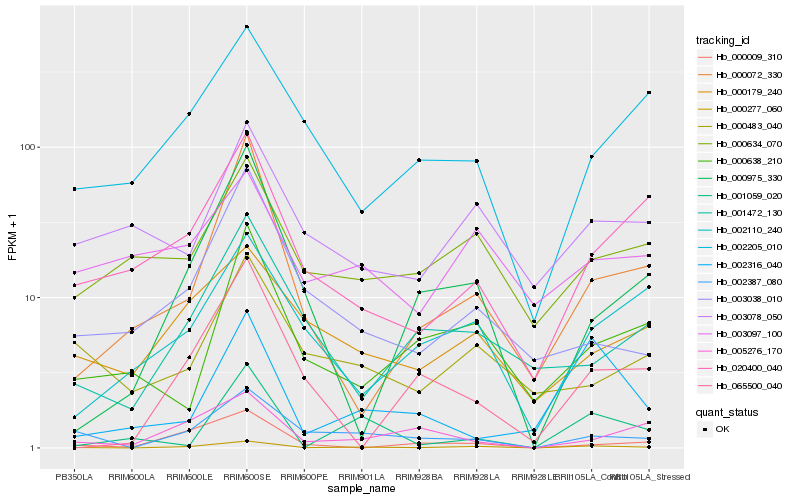
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000277_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 2 |
Hb_002387_080 |
0.2305583178 |
- |
- |
hypothetical protein JCGZ_25275 [Jatropha curcas] |
| 3 |
Hb_000072_330 |
0.2759498341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_020400_040 |
0.2802495983 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 5 |
Hb_000179_240 |
0.2817632771 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002110_240 |
0.2821145619 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 7 |
Hb_000634_070 |
0.2832996201 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 8 |
Hb_003038_010 |
0.2875498351 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_003078_050 |
0.2891275424 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 10 |
Hb_000638_210 |
0.2891577282 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000975_330 |
0.2904845329 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 12 |
Hb_000483_040 |
0.2905288912 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 13 |
Hb_001472_130 |
0.2915687952 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 14 |
Hb_002205_010 |
0.2940634275 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 15 |
Hb_065500_040 |
0.2953397693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000009_310 |
0.2958663484 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_002316_040 |
0.2992966353 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 18 |
Hb_001059_020 |
0.3005058125 |
- |
- |
PREDICTED: FBD-associated F-box protein At4g10400-like [Malus domestica] |
| 19 |
Hb_003097_100 |
0.3016020438 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 20 |
Hb_005276_170 |
0.3018721716 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |