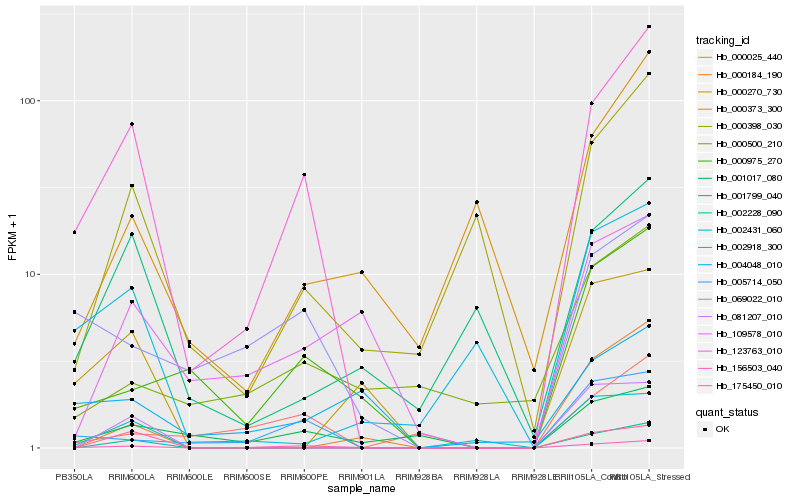
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123763_010 |
0.0 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 2 |
Hb_156503_040 |
0.1918576313 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_005714_050 |
0.2474827363 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 4 |
Hb_002228_090 |
0.2568179909 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 5 |
Hb_069022_010 |
0.2582323003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000398_030 |
0.259319606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000975_270 |
0.2622631303 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 8 |
Hb_000025_440 |
0.2788377073 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 9 |
Hb_001017_080 |
0.278918102 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 10 |
Hb_001799_040 |
0.281421079 |
- |
- |
- |
| 11 |
Hb_002918_300 |
0.2914169851 |
- |
- |
- |
| 12 |
Hb_175450_010 |
0.2975306198 |
- |
- |
- |
| 13 |
Hb_000270_730 |
0.2987491684 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 14 |
Hb_109578_010 |
0.2988979634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_081207_010 |
0.3040689471 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 16 |
Hb_000373_300 |
0.3057127494 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 17 |
Hb_004048_010 |
0.3058166927 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 18 |
Hb_002431_060 |
0.3078433502 |
- |
- |
- |
| 19 |
Hb_000500_210 |
0.3135538955 |
- |
- |
- |
| 20 |
Hb_000184_190 |
0.3140495911 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |