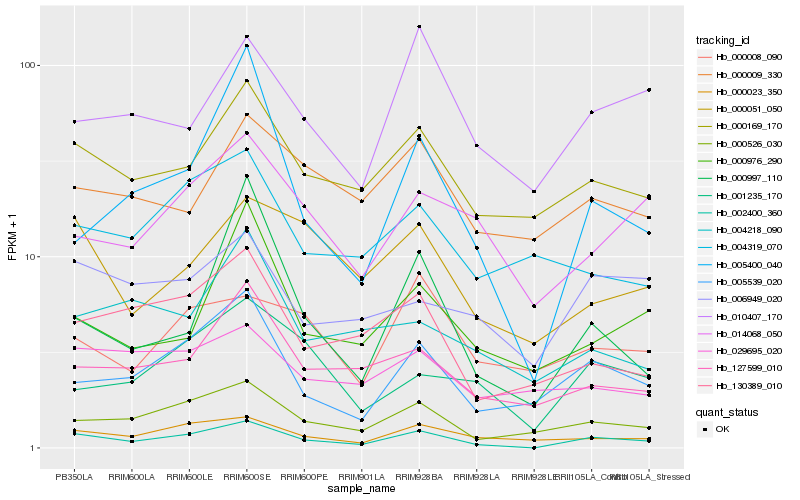
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_360 |
0.0 |
- |
- |
heparanase, putative [Ricinus communis] |
| 2 |
Hb_000526_030 |
0.175333898 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 3 |
Hb_005539_020 |
0.1775330195 |
- |
- |
- |
| 4 |
Hb_000169_170 |
0.1848717835 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 5 |
Hb_000023_350 |
0.1892198152 |
- |
- |
PREDICTED: probable folate-biopterin transporter 6 isoform X2 [Populus euphratica] |
| 6 |
Hb_127599_010 |
0.2018581276 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 7 |
Hb_130389_010 |
0.2030988045 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 8 |
Hb_000051_050 |
0.2066894337 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 9 |
Hb_005400_040 |
0.207194919 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 10 |
Hb_001235_170 |
0.2080230435 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 11 |
Hb_010407_170 |
0.2101981998 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 12 |
Hb_014068_050 |
0.2114201274 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 13 |
Hb_006949_020 |
0.2132314436 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine-C(5))-methyltransferase isoform X1 [Jatropha curcas] |
| 14 |
Hb_000976_290 |
0.2193825091 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 15 |
Hb_000008_090 |
0.2215451666 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 16 |
Hb_000009_330 |
0.2218169286 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_000997_110 |
0.2231125355 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 18 |
Hb_004319_070 |
0.2251382791 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 19 |
Hb_029695_020 |
0.2269094223 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004218_090 |
0.2278064591 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |