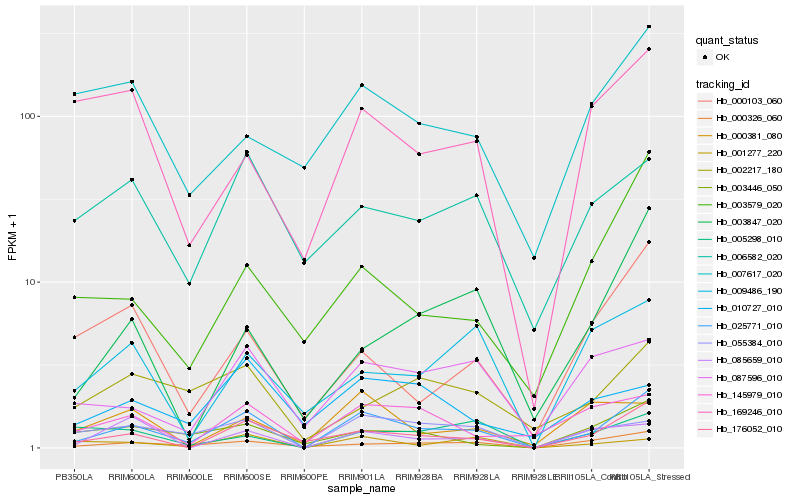
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025771_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 2 |
Hb_176052_010 |
0.1894535824 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_000326_060 |
0.2174005084 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 4 |
Hb_055384_010 |
0.2301834486 |
- |
- |
- |
| 5 |
Hb_087596_010 |
0.2422160195 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 6 |
Hb_003446_050 |
0.2467442663 |
- |
- |
hypothetical protein B456_002G034000, partial [Gossypium raimondii] |
| 7 |
Hb_145979_010 |
0.2486393549 |
- |
- |
PREDICTED: uncharacterized protein LOC105172316 [Sesamum indicum] |
| 8 |
Hb_002217_180 |
0.2576657306 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |
| 9 |
Hb_003579_020 |
0.2577586027 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 10 |
Hb_001277_220 |
0.260039651 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 11 |
Hb_006582_020 |
0.2604448611 |
- |
- |
Zinc finger protein ZFPM1 [Theobroma cacao] |
| 12 |
Hb_085659_010 |
0.2624564097 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 13 |
Hb_003847_020 |
0.265913933 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000103_060 |
0.2677504026 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 15 |
Hb_010727_010 |
0.2688851434 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 16 |
Hb_169246_010 |
0.270664841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009486_190 |
0.2711739378 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000381_080 |
0.2714019714 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair protein RAD16 [Jatropha curcas] |
| 19 |
Hb_005298_010 |
0.2728378199 |
- |
- |
- |
| 20 |
Hb_007617_020 |
0.2740421271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |