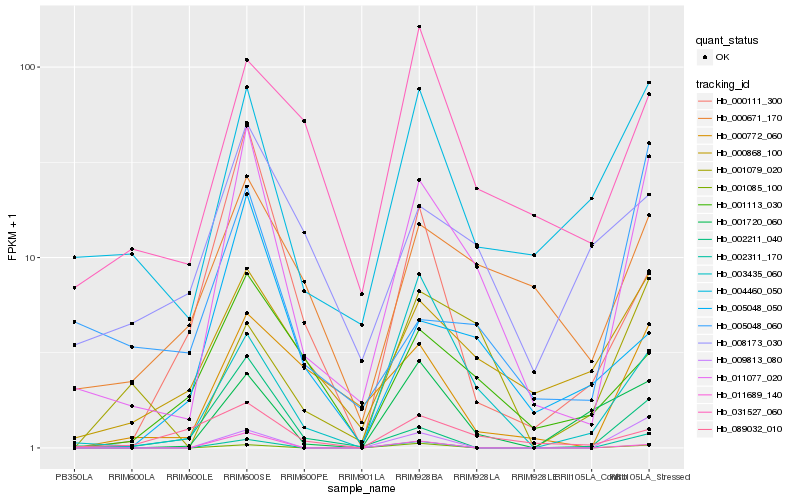
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011077_020 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_004460_050 |
0.2512379578 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 3 |
Hb_000868_100 |
0.251464264 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 4 |
Hb_000772_060 |
0.2665558206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000671_170 |
0.2736154595 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_002211_040 |
0.2885574635 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 7 |
Hb_001113_030 |
0.2896825364 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 8 |
Hb_001720_060 |
0.2953029425 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 9 |
Hb_002311_170 |
0.2969277579 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Eucalyptus grandis] |
| 10 |
Hb_001079_020 |
0.2988654741 |
- |
- |
hypothetical protein POPTR_0012s13720g [Populus trichocarpa] |
| 11 |
Hb_009813_080 |
0.3037704471 |
- |
- |
hypothetical protein CISIN_1g045991mg [Citrus sinensis] |
| 12 |
Hb_003435_060 |
0.304126885 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 13 |
Hb_001085_100 |
0.3053028565 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_008173_030 |
0.3059876728 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 15 |
Hb_005048_050 |
0.3062603802 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_000111_300 |
0.3064274504 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 17 |
Hb_031527_060 |
0.3066656445 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 18 |
Hb_089032_010 |
0.3080499755 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 19 |
Hb_005048_060 |
0.3081904861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011689_140 |
0.3084169345 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 4 [Jatropha curcas] |