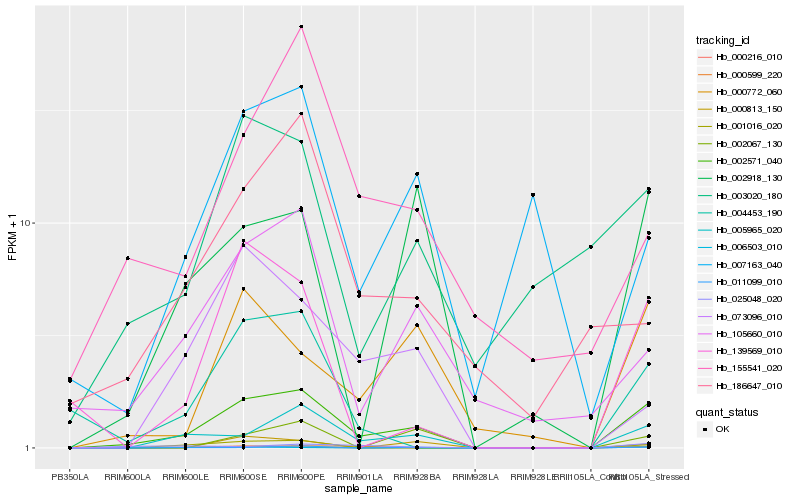
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002571_040 |
0.0 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 2 |
Hb_005965_020 |
0.2168243397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000216_010 |
0.2375009802 |
- |
- |
carbon monoxide dehydrogenase [Streptomyces sp. CNQ-509] |
| 4 |
Hb_000772_060 |
0.2691617929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002918_130 |
0.280329465 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 6 |
Hb_011099_010 |
0.2836805012 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_006503_010 |
0.2839864666 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 8 |
Hb_105660_010 |
0.285598873 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 9 |
Hb_004453_190 |
0.2942389195 |
- |
- |
- |
| 10 |
Hb_003020_180 |
0.3006708298 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 11 |
Hb_139569_010 |
0.3067036228 |
- |
- |
hypothetical protein JCGZ_24635 [Jatropha curcas] |
| 12 |
Hb_155541_020 |
0.3134277643 |
- |
- |
- |
| 13 |
Hb_073096_010 |
0.3136474015 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 45-like [Jatropha curcas] |
| 14 |
Hb_025048_020 |
0.3141543509 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_000813_150 |
0.3175756171 |
- |
- |
- |
| 16 |
Hb_001016_020 |
0.3185931591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007163_040 |
0.3190794694 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 18 |
Hb_002067_130 |
0.319502194 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 19 |
Hb_000599_220 |
0.3217060926 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 20 |
Hb_186647_010 |
0.3237264641 |
- |
- |
- |