| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
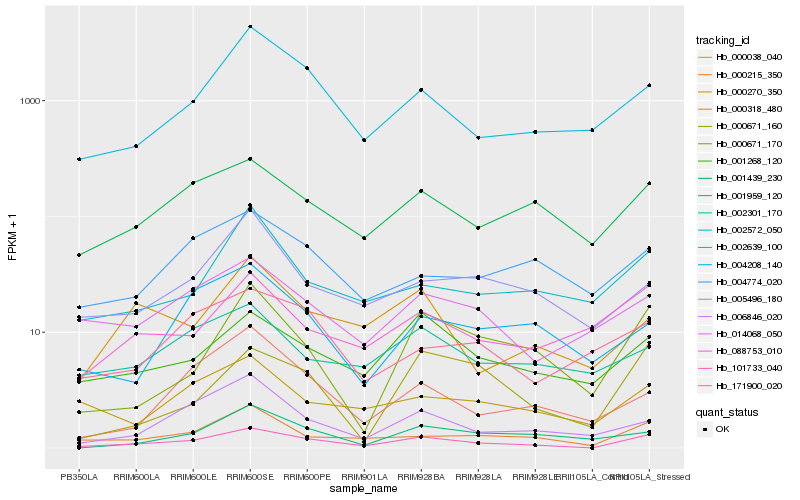
Hb_101733_040 |
0.0 |
- |
- |
cytochrome P450 CYP71B63v2 [Populus trichocarpa] |
| 2 |
Hb_000215_350 |
0.188413042 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_002639_100 |
0.2003047017 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 4 |
Hb_001268_120 |
0.2047335782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000671_170 |
0.2085294598 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_001439_230 |
0.2091928261 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006846_020 |
0.213642533 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 8 |
Hb_000671_160 |
0.2177296892 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 9 |
Hb_014068_050 |
0.2243102515 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 10 |
Hb_000038_040 |
0.2256883209 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 11 |
Hb_000318_480 |
0.22730273 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 12 |
Hb_088753_010 |
0.2289986135 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 13 |
Hb_002572_050 |
0.2297500653 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_005496_180 |
0.2307321041 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 15 |
Hb_004208_140 |
0.2311801631 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 16 |
Hb_000270_350 |
0.2322309056 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 17 |
Hb_171900_020 |
0.2345666794 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 18 |
Hb_004774_020 |
0.2346695716 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 19 |
Hb_001959_120 |
0.2360361403 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 20 |
Hb_002301_170 |
0.2368549839 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |