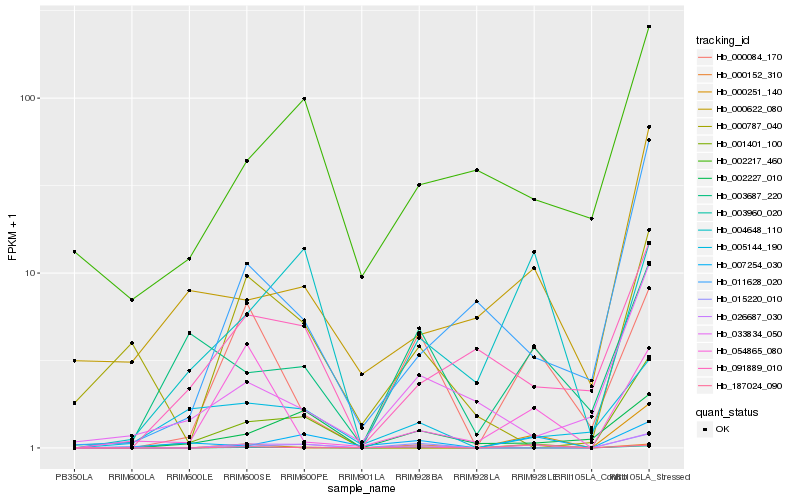
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026687_030 |
0.0 |
- |
- |
hypothetical protein AMTR_s00038p00220080 [Amborella trichopoda] |
| 2 |
Hb_015220_010 |
0.300606162 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 3 |
Hb_003960_020 |
0.3006918839 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_007254_030 |
0.3088717031 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_001401_100 |
0.3123646038 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 6 |
Hb_000084_170 |
0.3123931521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000787_040 |
0.3132392014 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 8 |
Hb_011628_020 |
0.3189613832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000622_080 |
0.3210797994 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 10 |
Hb_002217_460 |
0.3223240088 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 11 |
Hb_002227_010 |
0.329903277 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Populus euphratica] |
| 12 |
Hb_000251_140 |
0.3349134329 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 13 |
Hb_091889_010 |
0.3354089208 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000152_310 |
0.3354831509 |
- |
- |
- |
| 15 |
Hb_004648_110 |
0.3366620676 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_005144_190 |
0.3380164765 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 17 |
Hb_033834_050 |
0.3455487978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_054865_080 |
0.3489552601 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 19 |
Hb_003687_220 |
0.3494992538 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 20 |
Hb_187024_090 |
0.3529364607 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |