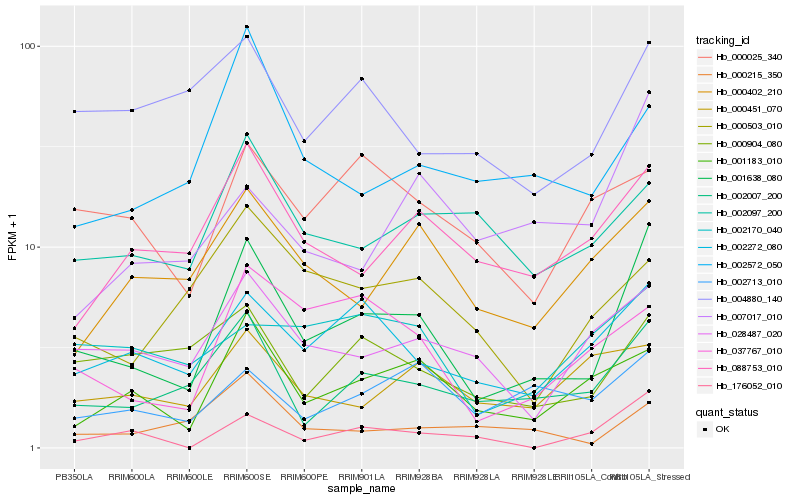
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 2 |
Hb_002007_200 |
0.1822345501 |
- |
- |
- |
| 3 |
Hb_002272_080 |
0.2018370361 |
- |
- |
- |
| 4 |
Hb_001183_010 |
0.2119829028 |
- |
- |
- |
| 5 |
Hb_028487_020 |
0.2164072035 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 6 |
Hb_088753_010 |
0.2166205678 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 7 |
Hb_037767_010 |
0.2204029501 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000904_080 |
0.2307292676 |
- |
- |
alpha-amylase-like protein [Vitis labrusca x Vitis vinifera] |
| 9 |
Hb_000402_210 |
0.2312984991 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |
| 10 |
Hb_004880_140 |
0.2340981554 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 11 |
Hb_002572_050 |
0.2348474903 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_002713_010 |
0.2361096666 |
- |
- |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 13 |
Hb_002097_200 |
0.2381308084 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 14 |
Hb_000215_350 |
0.2407721715 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_176052_010 |
0.2411788355 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_007017_010 |
0.2444451821 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 17 |
Hb_000503_010 |
0.2448289733 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 18 |
Hb_002170_040 |
0.2478481182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000025_340 |
0.2488230701 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 20 |
Hb_000451_070 |
0.2489721722 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |