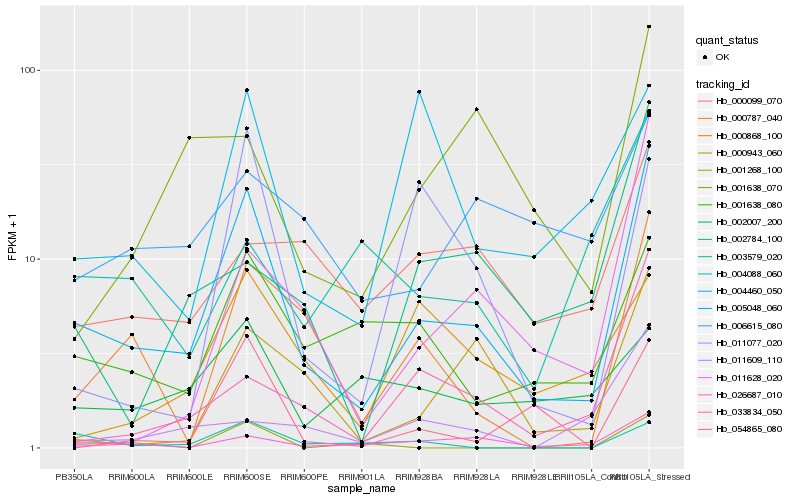
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005048_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_026687_010 |
0.2423588454 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 3 |
Hb_000787_040 |
0.2505804267 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 4 |
Hb_001638_080 |
0.2662700924 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 5 |
Hb_033834_050 |
0.2779000393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000943_060 |
0.2804410283 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 7 |
Hb_011628_020 |
0.2919673103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_054865_080 |
0.2923173087 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 9 |
Hb_002784_100 |
0.2932146586 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 10 |
Hb_004088_060 |
0.2982016819 |
- |
- |
hypothetical protein POPTR_0012s01875g [Populus trichocarpa] |
| 11 |
Hb_001638_070 |
0.3062182284 |
transcription factor |
TF Family: bHLH |
Basic helix-loop-helix DNA-binding family protein [Theobroma cacao] |
| 12 |
Hb_000099_070 |
0.3064303787 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 13 |
Hb_011077_020 |
0.3081904861 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000868_100 |
0.3160814739 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 15 |
Hb_006615_080 |
0.316848075 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 16 |
Hb_011609_110 |
0.3170472701 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 17 |
Hb_003579_020 |
0.3176494082 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 18 |
Hb_004460_050 |
0.3184629159 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 19 |
Hb_001268_100 |
0.3200963453 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 20 |
Hb_002007_200 |
0.3217299821 |
- |
- |
- |